Abstract
Sonic hedgehog (Shh) is a signaling molecule that is important for defining patterning in the developing vertebrate central nervous system. After translation, Shh autoproteolyzes and covalently attaches cholesterol to the newly formed carboxyl terminus, a modification crucial for normal Shh signaling. Presented here is evidence that acute severe sterol deprivation in cultured Chinese hamster ovary cells expressing mouse Shh (mShh) inhibits autoprocessing of the protein. These conditions allowed the first detailed kinetic analysis of mShh autoprocessing and turnover rates revealing that cells rapidly degrade both precursor and mature mShh regardless of sterol content and sterol deprivation increases the rate of precursor degradation. Inhibition of mShh autoprocessing also allowed the determination of the subcellular localization of mShh precursor which accumulates in a pre-medial Golgi intracellular compartment. Finally, the precursor form of mShh that results from autoprocessing inhibition appears to accumulate as an amide rather than a stable thioester.
The hedgehogs (hh) are a family of genes that are responsible for pattern formation in the developing embryo (1). A member of this family, sonic hedgehog (Shh), is expressed in the developing central nervous system of vertebrate embryos where it functions in multiple patterning events including induction of the floor plate and the differentiation of specific neurons types (2). Shh also has other functions that include controlling the differentiation of skeletal and muscular tissue (3) and coordinating the anterior-posterior patterning of the developing limb (4).
When expressed in its target tissues, Shh exhibits a tightly controlled localization that is dependent on lipid modification (5–7). Shh is produced as a 47–49 kDa-secreted (depending on species) holoprotein that posttranslationally cleaves to give two mature proteins: a 19-kDa amino-terminal fragment that remains cell associated and a 29–31-kDa carboxy terminal fragment that is released from the cell (5, 8, 9). The membrane-associated amino-terminal fragment contains the signaling portion of the molecule. The mouse Shh (mShh) precursor carboxy terminus encodes the autoprocessing domain which acts only in cis (10). Beachy et al. (11) has demonstrated that the autoproteolysis occurs concomitantly with the covalent attachment of cholesterol to the nascent carboxy terminus of the 19-kDa signaling fragment. Mechanistically, this proteolysis proceeds through two steps. First, an internal thioester is formed by attack of the sulfhydryl of the Cys in a conserved Gly-Cys-Phe motif on the carboxamide of the immediately amino-terminal Gly. Next, the hydroxyl group of cholesterol attacks the intermediate thioester to simultaneously eject the carboxy-terminal protein fragment and form the cholesterol ester at the new carboxy terminus. In vitro, the rate of the processing reaction of a truncate of Drosophila hh is accelerated by the addition of some sterols (11, 12). To date, the cholesterol linkage to hedgehog proteins remains the sole example of this lipid modification.
Several groups have hypothesized that cholesterol deficiency may prevent normal hedgehog posttranslational processing (1, 11, 13). Recent studies have found that transient treatment of cultured cells expressing Shh with inhibitors of cholesterol biosynthesis did not affect Shh autoprocessing, a result that has also been echoed in this laboratory (12, 14). In the current study, a Chinese hamster ovary 7 (CHO-7) cell line stably and constitutively expressing an epitope-tagged version of mShh was subjected to transient severe sterol depletion by using a combination of (i) inhibition of endogenous cholesterol synthesis with compactin, and (ii) equilibrium partition removal of plasma membrane sterols with cyclodextrin. These conditions prevented the posttranslational processing and secretion of mShh by inhibiting both steps of the processing reaction. This discovery allowed the first detailed kinetic analysis of the rates of mShh autoproteolysis and turnover. These studies revealed that both precursor and mature forms of mShh are turned over rapidly by cells and that the rate of precursor degradation is increased by sterol deprivation.
Materials and Methods
Materials.
Newborn calf lipoprotein-deficient serum (d > 1.215 g/ml), sodium mevolonate, sodium compactin, and the mouse IgG 2001 mAb were prepared as described (15–17). Methyl-β-cyclodextrin, monoclonal mouse α-FLAG(M2) antibody, and mouse α-FLAG(M2)⋅agarose were obtained from Sigma; 35S EasyTag Protein Express Label from New England Nuclear; Endo H and PNGase F glycosidases from New England Biolabs; Complete protease inhibitor tablets from Boehringer Mannheim; monoclonal mouse α-transferrin receptor antibody from Zymed; polyclonal donkey α-mouse⋅horseradish peroxidase antibody from The Jackson Laboratory; expression plasmid pCDNA3.1 (−) from Invitrogen; the Transformer mutagenesis system from CLONTECH; the Immunocatcher system from Cytosignal (Irvine, CA); and BioMax MS film and Personal LE intensifying screens from Kodak.
Recombinant Plasmids.
A vector encoding FLAG-epitope-tagged mouse sonic hedgehog (pCDNA-FLAG-mShh) was produced as follows: a PCR fragment containing codons 1–437 of mShh was amplified by using primers that included an amino-terminal EcoRI restriction site and Kozac sequence and a carboxy terminal HindIII site and subcloned into pCDNA3.1(−). The resulting plasmid was mutated by using the Transformer system to insert sequential ClaI and KpnI sites between codons 32 and 33 of the mShh gene. A synthetic double-stranded oligonucleotide encoding compatible cohesive ends and a double copy of the FLAG-epitope tag was inserted into the ClaI/KpnI site. The sequence and orientation of the gene was verified by DNA sequencing of both strands.
Cultured Cells.
The cell lines were all derived from CHO-7, a subline of CHO-K1 cells selected for growth in lipoprotein-deficient serum. Cells were maintained as a monolayer in medium A (a 1:1 mixture of Ham's F-12 medium and DMEM containing 10% (vol/vol) newborn calf lipoprotein-deficient serum/100 units/ml penicillin/100 μg/ml streptomycin sulfate) at 37°C in a 9% CO2 incubator. CHO/mShh cells and CHO/− control cells were isolated by transfection of CHO-7 cells with plasmids pCDNA-FLAG-mShh and pCDNA3.1(−), respectively, followed by selection for growth in 10% newborn calf lipoprotein-deficient serum and 750 μg/ml G418. G418-resistant colonies were cloned by limiting dilution and screened for expression by immunoblot analysis of Triton X-100 lysates (described below under Immunoblot analysis) with a polyclonal α-FLAG antibody. Stock cultures of CHO/mShh and CHO/− control cells were maintained in medium A supplemented with 500 μg/ml G418.
Detergent Lysis of Cultured Cells.
Cells were washed twice with room temperature Dulbecco's PBS and then scraped into PBS. Replicate dishes were pooled and centrifuged at 1,000 × g for 5 min at 4°C. The cell pellet was washed once with PBS, centrifuged, resuspended in 200 μl per dish of lysis buffer [1% (vol/vol) Triton X-100/50 mM Tris·HCl (pH 7.4)/150 mM NaCl/20 mM MgCl2/2 mM CaCl2/Complete protease inhibitors), and allowed to stand for 30 min at 4°C. The resulting suspension was centrifuged at 100,000 × g for 30 min at 4°C. The supernatant was either used immediately or frozen in liquid nitrogen and stored at −78°C.
Processing of mShh in Cultured Cells.
On day 0, cells were set up in monolayers at a density of 7 × 105 cells per 60-mm dish in medium A supplemented with 500 μg/ml G418, thereby allowing four dishes per experimental condition. On day 1, the dishes were split into two equal sets, one of which was treated with the radioactive label (experimental samples) whereas the other received nonradioactive amino acid supplements at the same concentrations used in the experimental samples (reserved samples). On day 1, at −1 h, the cells were refed with medium A supplemented with 500 μg/ml G418 or sterol-depriving medium B (medium A supplemented with 500 μg/ml G418/10 μM sodium compactin/250 μM sodium mevolonate/3.7 mM methyl-β-cyclodextrin). Sterol-depriving additives were included in all subsequent media for the appropriate samples. At 0 h, the cells were washed twice and refed with medium C [DMEM (high glucose, without Met) supplemented with 10% (vol/vol) newborn calf lipoprotein-deficient serum/100 units/ml penicillin/100 μg/ml streptomycin sulfate/500 μg/ml G418/17.25 μg/ml L-Pro]. At +0.5 h, the cells were washed once with medium C and then refed with medium C supplemented with 175 μCi/ml 35S EasyTag Protein Labeling mixture (or the equivalent amount of Met) and incubated for the indicated pulse time. The cells were then washed twice and refed with medium C supplemented with 86 μg/ml Met and 175 μg/ml Cys (5X normal concentrations) and incubated for the indicated chase time. The detergent lysates of all samples were prepared as described above. The detergent-soluble fractions from the experimental samples (two pooled dishes each sample) were mixed with 20 μl of preimmune whole rabbit serum/20 μg irrelevant mouse mAb IgG 2001/100 μl of Protein A/G⋅agarose beads and rotated at 4°C for 2 h. After rotation, the experimental samples were centrifuged at 1,000 × g for 5 min at 4°C. The resulting supernatant was mixed with 100 μl of protein A/G⋅agarose and rotated for 2 h at 4°C. At the same time, the detergent-soluble fractions from the reserved samples (two pooled dishes each sample) were mixed with 50 μl of mouse α-FLAG(M2)⋅agarose beads (prewashed three times with 500 μl of PBS) and rotated at 4°C for 4 h. Both sets were centrifuged at 1,000 × g for 5 min at 4°C. The supernatants from the experimental samples were mixed with the precoated beads from the reserved samples and rotated at 4°C for 2 h. The beads were then washed six times with 0.5 ml of wash buffer [1% (vol/vol) Triton X-100/50 mM Tris·HCl (pH 7.4)/800 mM NaCl/800 mM urea/20 mM MgCl2/2 mM CaCl2/Complete protease inhibitors) by using Immunocatcher spin columns. The washed beads were resuspended in 50 μl of 1X SDS loading buffer [75 mM Tris·HCl (pH 6.8)/2% SDS/10% glycerol/100 mM DTT/0.01% (wt/vol) bromophenol blue], allowed to stand for 15 min at room temperature, boiled for 5 min, and eluted into a new tube by centrifugation at 14,000 × g for 3 min at ambient temperature. The immunoprecipitated mShh proteins were separated by SDS/PAGE, the gel was dried, and then exposed for autoradiography by using the Personal LE screen/BioMax MS film system for the indicated time.
Immunoblot Analysis.
Samples, normalized to a total protein content of 50 μg per sample, were subjected to SDS/PAGE and transferred electrophoretically to Immobilon-P membranes. The membranes were blocked and blotted with primary and secondary antibodies according to the manufacturer's recommendations. The bound antibodies were visualized with the Supersignal Plus chemiluminescent horseradish peroxidase substrate using prestained molecular weight markers for calibration.
Glycosidase Treatment.
On day 0, cells were set up at 5 × 105 per 100-mm dish with one dish per experimental condition in medium A supplemented with 500 μg/ml G418. On day 3, cells were refed with medium A or B. At +4 h, cells were scraped into medium and centrifuged at 1,000 × g for 5 min at 4°C. The cell pellets were washed with 1 ml of ice-cold PBS and centrifuged. The cell pellets were resuspended in 400 μl per dish of lysis buffer [10 mM Hepes (pH 7.6)/1.5 mM MgCl2/10 mM KCl/250 mM sucrose/5 mM EGTA/5 mM EDTA/Complete protease inhibitors], passed 20 times through a 22-gauge needle, and centrifuged. The supernatant was transferred to a new tube and centrifuged at 4°C for 10 min at 15,000 × g. The resulting pellets were resuspended in 110 μl per dish of denaturation buffer (lysis buffer supplemented with 100 mM NaCl/0.5% SDS/1% βmercaptoethanol) and boiled for 10 min. The samples were allowed to cool to ambient, diluted with 11 μl of the manufacturer's 10X reaction buffer, and treated with 1,000 units of the indicated glycosidase (or an equivalent amount of 1X buffer) for 1 h at 37°C. The samples were mixed [4:1 (vol:vol)] with 5X SDS-loading buffer and boiled for 10 min. Samples, normalized to a total protein content of 50 μg per sample (for mShh gels) or 20 μg per sample (for TfR gels) were subjected to SDS/PAGE. Proteins were transferred to membranes and immunoblotted as described above.
Results
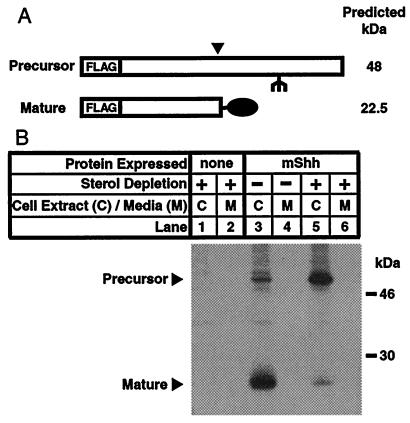
Initial attempts to inhibit processing of mShh by using inhibitors of endogenous cholesterol synthesis such as compactin, zaragosic acid, and AY9944, were unsuccessful. However, application of recently developed techniques for severely starving cells of sterols (18) induced a dramatic effect on hedgehog processing as revealed by pulse-chase studies. When CHO/mShh cells are grown in normal lipoprotein-deficient media, the freshly synthesized mShh protein has almost completely converted to the mature form by 30 min after synthesis (Fig. 1, lanes 3 and 4). Treatment of the cells with sterol-depriving conditions blocked autoprocessing and led to a decrease in the amount of mature hedgehog and a corresponding accumulation of the precursor form (Fig. 1, lanes 5 and 6). In both control and treated cells, the mature and precursor forms of mShh are retained in the cells. This remains the case if the chase time is extended to 4 h with no mShh being detected in the media at any time. Control experiments in which the immunoprecipitation media was mixed with detergent lysates from CHO/mShh cells indicated that both forms of mShh could be efficiently immunoprecipitated from the media when present. Therefore, the absence of mShh in the media was not caused by an inability to immunoprecipitate the protein from the media.
Figure 1.
Inhibition of mShh autoprocessing by sterol deprivation. (A) Schematic representation of the precursor and mature forms of recombinant mShh protein. FLAG indicates the double copy of the FLAG-epitope tag. The inverted filled triangle indicates the site of autoproteolytic cleavage, and the filled oval indicates the site of concomitant attachment of cholesterol. The fork indicates a site for N-linked glycosylation. (B) Pulse-chase experiment demonstrating that sterol deprivation inhibits mShh processing and mShh remains cell associated. CHO-7 cells, where indicated stably expressing mShh, were grown overnight in medium A. One hour before metabolic labeling, the medium was replaced, as indicated, with sterol-depriving medium (see Materials and Methods). Cells were subjected to a 30-min pulse labeling with 35S-labeled Met and 30-min chase. Recombinant mShh was immunoprecipitated from detergent lysates or media with mouse α-FLAG(M2)⋅agarose and subjected to SDS/PAGE (12% gel).
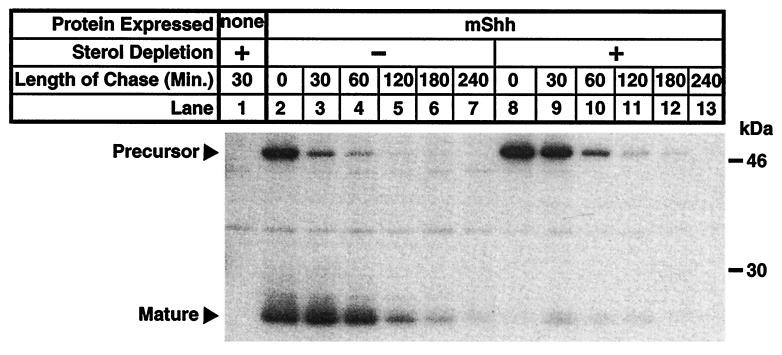
The next study examined the dynamics of mShh autoprocessing and degradation by pulse-chase experiments with CHO/mShh cells. When cells are grown in normal lipoprotein-deficient media, the newly synthesized precursor converts rapidly to mature form which is then completely degraded within 4 h (Fig. 2, lanes 2–7). However, when cells are treated with sterol-depriving conditions, the conversion of precursor to mature form is blocked and the resulting arrested precursor turns over within 2 h (Fig. 2, lanes 8–13). Turnover of precursor, conversion of precursor to mature form, and degradation of mature form all exhibit pseudo first-order kinetics with apparent rate constants of 0.030/min−1 for precursor degradation in sterol-deprived cells, 0.038/min−1 for the disappearance of precursor in untreated cells, and 0.019/min−1 for mature-form degradation in untreated cells. These relative rates correspond to half-lives of 23, 18, and 37 min, respectively. Assuming the processing reaction is first order and extrapolating the half-life of cleavage in untreated cells from the ratio of precursor to mature forms at the beginning of the chase period predicts a half-life of 18 min for the processing reaction.
Figure 2.
Pulse-chase experiment showing the time course of mShh autoprocessing and turnover of precursor and mature forms of mShh in normal and sterol-deprived cells. CHO-7 cells, where indicated stably expressing mShh, were grown overnight in medium A. One hour before metabolic labeling, the medium was replaced, as indicated, with sterol-depriving medium. Cells were labeled for 30 min with 35S-labeled Met followed by a variable-length chase (see Materials and Methods). Recombinant mShh was immunoprecipitated from detergent lysates of cell pellets with mouse α-FLAG(M2)⋅agarose and subjected to SDS/PAGE (12% gel).
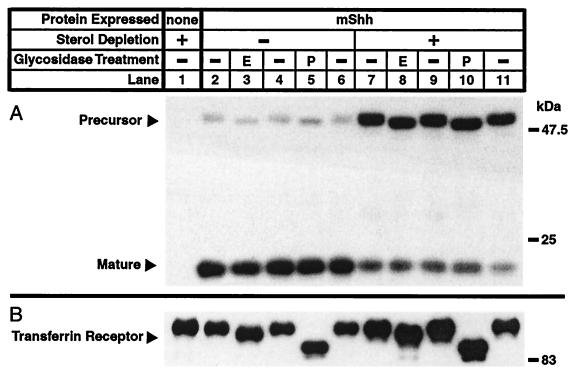
The observation that both forms of mShh remain cell associated prompted study of the subcellular localization of mShh in CHO/mShh cells with glycosidase-sensitivity experiments. In untreated cells, the bulk of the steady-state accumulation of mShh protein is in the mature form (Fig. 3A, lanes 2–6). The apparent molecular weight of the glycosylated precursor in these cells is sensitive to the actions of EndoH (lane 3) which cleaves only high mannose carbohydrates and PNGaseF (lane 5) which cleaves all N-linked sugars. In CHO/mShh cells that have been sterol deprived for 4 h before harvest, the steady-state partition between the forms of mShh has shifted dramatically to favor the precursor (Fig. 3A, lanes 7–11). However, the precursor form is still sensitive to both glycosidases (lanes 8 and 10). Therefore, in both treated and untreated cells, the precursor never proceeds past the cis-Golgi compartment of the secretory pathway. The secreted carboxy terminus, which contains the glycosidation site, has previously been shown to be resistant to EndoH but sensitive to PNGaseF (19). The Endo H sensitivity of the transferrin receptor, a protein that proceeds through the secretory pathway after synthesis (20), is not perturbed by sterol deprivation (Fig. 3B).
Figure 3.
Glycosidase-sensitivity experiment demonstrating the glycosylation state of mShh precursor in normal and sterol-deprived cells. (A) Effects of sterol deprivation on the subcellular localization of mShh and the steady-state partition between precursor and mature forms of mShh. Treatment of samples with EndoH is indicated with an “E” whereas treatment of samples with PNGase F is indicated with a “P”. (B) Effects of sterol deprivation on the subcellular localization of the transferrin receptor (TfR). CHO-7 cells, where indicated stably expressing mShh, were grown for 48 h in medium A. Four hours before harvest, the media was replaced, as indicated, with sterol-depriving medium. At harvest, membrane fractions were prepared, denatured, and incubated in the presence or absence of PNGase F or Endo H (see Materials and Methods). The samples were then subjected to SDS/PAGE (9% for mShh; 6.5% for TfR control), transferred to Immobilon-P, and immunoblotted with either mouse α-FLAG(M2) or mouse α-TfR primary antibodies and donkey α-mouse⋅horseradish peroxidase secondary antibody. Blots were developed by using a chemiluminescent substrate for horseradish peroxidase.
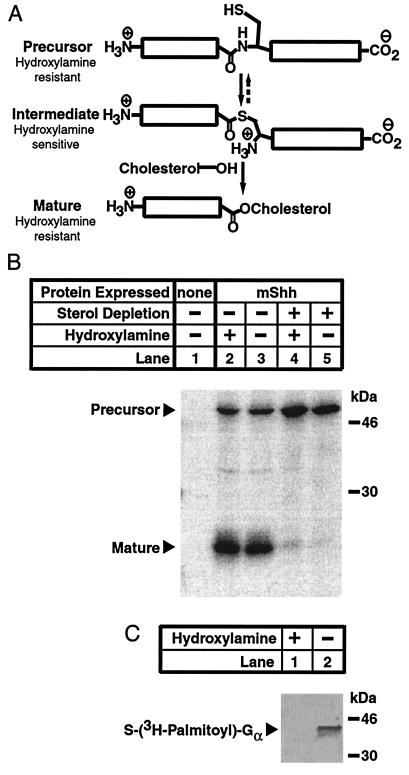
A metastable apparent precursor form of mShh could accumulate in two states in sterol-deprived CHO/mShh cells (i) as the amide precursor, or (ii) as the thioester intermediate (Fig. 4A). These states are indistinguishable by apparent molecular weight. Owing to the unique chemical properties of thioesters, they are cleaved by neutral hydroxylamine whereas esters and amides are not (Fig. 4A) (21). Thus, treatment of the apparent precursor form of mShh with hydroxylamine could distinguish between these two states. When the mShh proteins, pulse labeled and immunoprecipitated from untreated cells before the completion of processing, are treated with hydroxylamine, the precursor form is resistant to cleavage (Fig. 4B, lanes 2 and 3). The complete absence of a carboxy terminal band (30 kDa), which would be readily apparent on cleavage of the intermediate form, implies that none of the apparently mature form is derived from chemically induced cleavage. Likewise, when the precursor mShh, isolated from sterol-deprived cells in the same manner, is treated with hydroxylamine, no cleavage of precursor is induced (Fig. 4B, lanes 4 and 5). As a control, in vitro-translated G protein α, labeled with a tritiated palmitoyl thioester (22), was mixed with unlabeled lysates from sterol-deprived CHO/mShh cells and treated with hydroxylamine which completely removed the palmitoyl group (Fig. 4C). Thus, sterol deprivation does not produce a factor that stabilizes thioesters.
Figure 4.
Resistance of mShh precursor to neutral hydroxylamine. (A) Schematic representation of the chemical reactions carried out during mShh autoprocessing. First, mShh precursor converts to a semistable thioester intermediate by attack of the side chain thiol of Cys-199 on the carboxamide of the immediately amino-terminal Gly. Next, the intermediate form of mShh is converted to the cholesterol ester mature form by the attack of cholesterol on the thioester. Amides and esters are resistant to cleavage by neutral hydroxylamine whereas thioesters are labile to such cleavage. (B) Resistance of mShh precursor to neutral hydoxylamine. CHO-7 cells, where indicated stably expressing mShh, were grown overnight in medium A. One hour before metabolic labeling, the medium was replaced, as indicated, with sterol-depriving medium. Cells were labeled for 30 min with 35S-labeled Met followed by a 15-min chase. Recombinant mShh was immunoprecipitated from detergent lysates with mouse α-FLAG(M2)⋅agarose (see Materials and Methods). Before elution from the beads, immunoprecipitated mShh proteins were treated, as indicated, with either 1 M Tris·HCl (pH 7.4) or 1 M hydroxylamine (pH 7.4) for 30 min at 37°C. The resulting protein mixtures were eluted as described and subjected to SDS/PAGE (12% gel). (C) Lability of the palmitate thioester of G protein α to neutral hydroxylamine. Control immunoprecipitates were prepared from cells treated as described above without radiolabeling. Immediately before hydroxylamine treatment, the samples were treated with in vitro-translated G protein α that had been labeled with tritiated palmitate (24,000 cpm per sample). Before elution from the beads, these samples were then subjected, as indicated, to treatment with either 1 M Tris·HCl (pH 7.4) or 1 M hydroxylamine (pH 7.4) for 30 min at 37°C. The resulting protein mixtures were eluted from the beads, subjected to SDS/PAGE (12% gel), and transferred electrophoretically to an Immobilon P membrane.
Discussion
Inhibition of Shh Autoprocessing.
Transiently blocking the endogenous synthesis of cholesterol did not perturb normal hedgehog posttranslational processing. In cultured cells that are unable to produce cholesterol, complete depletion of cholesterol is achieved by dilution into the membranes of daughter cells after approximately three passages in cholesterol-deficient media (23, 24). Therefore, it seemed likely that the lack of apparent effect on mShh processing by inhibitors of cholesterol synthesis was caused by a failure to sufficiently lower cellular cholesterol levels within the limited time span of the experiments. Acutely and severely lowering the sterol levels of the cells by using a combination of physically removing existing cellular sterols with the sterol-complexing agent methyl-β-cyclodextrin and inhibiting cholesterol synthesis with the hydroxymethyl glutaryl CoA reductase inhibitor compactin (18) completely blocked processing of the mShh precursor. Although this effect is probably caused by cholesterol starvation, it is impossible to rule out removal of other regulatory sterols by this process.
When mShh processing is blocked in this manner, the precursor form builds up proportionally within the cells but does not accumulate to very high levels. The accumulated precursor remains associated with the cells expressing the protein rather than being secreted into the surrounding media. One possible explanation for this failure to secrete hh could be blockade of all protein transport. However, the same sterol-depleting conditions fail to block the normal transport of the transferrin receptor to the plasma membrane, implying that bulk protein trafficking is not inhibited. Another potential explanation is that mShh precursor synthesized in the absence of cholesterol does not fold properly and is retained in the endoplasmic reticulum by the protein quality control system (25). However, a mutant mShh (mShhC198A) that is unable to process is retained within cells with normal levels of cholesterol (unpublished observations). In as much as Cys-198 is mechanistically required to be both conformationally flexible and solvent exposed, it seems unlikely that this mutation would result in gross misfolding of mShh. The more likely explanation is therefore that the cell actively retains the precursor form of mShh within the secretory pathway. Glycosidase-sensitivity experiments discussed below suggest that the accumulated precursor does not cycle back from the plasma membrane.
Rates of Shh Processing and Degradation.
One must always exert caution in assuming that the kinetic behavior of proteins in transfected cells is similar to that in endogenous systems. To minimize experiment-to-experiment variability and perturbations induced by extreme overexpression of mShh, the cells used in this study were produced by stable transfection of the mShh gene and selected for moderate protein expression levels (CHO/mShh cells). In these cells, the bulk of the mShh protein detectable by immunoblot (>95% by densitometry) has processed to the mature form as was the case in prior studies in other laboratories (19, 26).
Pulse-chase experiments allowed an examination of the kinetics of the processing reaction (Fig. 2). In untreated CHO/mShh cells, the rate of precursor disappearance is dominated by the kinetics of the processing reaction rather than by precursor degradation. The processing is an apparent first-order reaction with t1/2 of 18 min (kobs). Thus, as would be expected from the immunoblotting results, mShh precursor rapidly processes to the mature form. In sterol-deprived cells, the processing reaction is completely suppressed and precursor disappears through degradation with first-order kinetics and an observed t1/2 of 23 min (kdegrad). In untreated cells, the rate of disappearance of the precursor form of mShh (kobs) is related to the rates of processing and degradation by the equation: kobs = kdegrad + kprocess. Substitution of the kobs (from untreated cells) and kdegrad (from sterol-deprived cells) gives kprocess = 0.0084 or an expected t1/2 of 83 min for the processing reaction in untreated cells. This is clearly at odds with the observed half-life of processing (18 min) in untreated cells. This finding suggests that sterol deprivation has two effects: (i) blocking the processing reaction, and (ii) increasing the degradation rate of the precursor form of mShh.
Several interesting conclusions arise from this kinetic analysis. First, the levels of both forms of mShh are tightly regulated by protein degradation with half-lives of <1 h. Such rapid proteolytic degradation of mShh protein might allow the attenuation of extracellular signaling when it is not contextually appropriate. Second, the normal rate of processing is rapid enough that the bulk of mShh in a given cell will normally exist as the mature form. These kinetic effects are mirrored in the bulk distribution of the hh expressed within normal cells where almost all (>95%) of the mShh detected by immunoblotting is in the mature form. In sum, these results suggest a model where the freshly synthesized mShh in untreated cultured cells is committed to the processing pathway.
Localization of Shh.
The observation that both forms of mShh remain cell associated prompted an exploration of the subcellular localization of the precursor form of mShh. Glycosidation-sensitivity studies revealed that the precursor fragment is held in a pre-medial Golgi compartment regardless of the sterol content of the cells. Control experiments revealed that sterol deprivation does not perturb bulk protein secretion, a result that mirrors the prior work of Simons et al. (18, 27). Whereas these localization results could arise artifactually from an abnormal glycoform that is always EndoH sensitive, the N-linked sugar on the normally secreted carboxy-terminal fragment of mShh has been found resistant to Endo H (19). Thus, the precursor form of mShh observed after sterol depletion of CHO/mShh cells has not recycled from the plasma membrane. This finding suggests that cultured cells expressing mShh have a mechanism for internal retention of the precursor form. Whereas this result could be caused by specific mistrafficking of mShh induced by sterol deprivation, a point mutant (mShhC198A) that is mechanistically unable to process is also held in a preGolgi compartment (results not shown). These findings imply that progression further into the secretory pathway requires autoprocessing of mShh.
Mechanism of Shh-Processing Arrest.
The realization that a precursor form of mShh does accumulate in cells that are sterol depleted led to an exploration of the mechanism of processing arrest (Fig. 4). Beachy and colleagues have previously demonstrated that a heterologously expressed truncate of Drosophila hh that encodes the processing domain will undergo autoprocessing in vitro on treatment with small nucleophiles such as DTT and hydroxylamine. This finding could be explained by one of two models. In one model (stepwise), the hh precursor converts to a metastable thioester intermediate after translation and this intermediate is cleaved by addition of nucleophile. Alternatively, there could be an equilibrium established between precursor and intermediate forms of hh with the reactive thioester being trapped by the nucleophile and thus driving the equilibrium toward the thioester (dynamic model). When freshly synthesized precursor was isolated from either untreated or sterol-deprived CHO/mShh cells, the protein was resistant to hydroxylamine under conditions that resulted in cleavage of known thioesters (28). Whereas such an experiment cannot definitively rule out the presence of a stable thioester intermediate, it does clearly favor the dynamic model of hh processing in mammalian cells. It may be the case that sterol deprivation regulates, either kinetically or thermodynamically, the formation of the thioester intermediate. However, the conditions of protein purification used in this study are likely to disrupt the processing domain structure and thus the potential equilibrium between precursor and intermediate forms. Therefore, the precise mechanism of regulation of mShh autoprocessing remains elusive.
Conclusions
These studies demonstrate that the autoprocessing of mShh protein in cultured mammalian cells can be inhibited by acute sterol deprivation with sterol-deprived cells being unable to produce a mature Shh signaling molecule. The extreme conditions required to achieve this inhibition suggest that this mechanism of autoprocessing inhibition is not likely to be relevant to mShh pathophysiology. However, these conditions did allow the first studies of the rates of mShh autoprocessing and turnover as well as the determination of the subcellular localization of the precursor form. It is apparent that in cultured cells, the rates of both autoprocessing and protein turnover are extremely rapid. Furthermore, the location of autoprocessing is intracellular and most probably within a secretory pathway compartment before the medial Golgi apparatus.
Acknowledgments
The author thanks Michael Brown and Joseph Goldstein for their invaluable advice and support; Alex Duncan for his gift of G protein; Andrew McMahon for the plasmid-encoding mShh; Rob Rawson, Axel Nortruft, and Elizabeth Duncan for helpful discussions; and Gloria Brunshende, Debbie Morgan, and Rita Wilson for technical assistance. National Institutes of Health Grant HL20948 and a Helen Hay Whitney Foundation postdoctoral fellowship supported this research.
Abbreviations
- CHO
Chinese hamster ovary
- hh
hedgehog
- Shh
sonic hedgehog
- mShh
mouse Shh
- d
density
- TfR
transferrin receptor
References
- 1.Hammerschmidt M, Brook A, McMahon A P. Trends Genet. 1997;13:14–21. doi: 10.1016/s0168-9525(96)10051-2. [DOI] [PubMed] [Google Scholar]
- 2.Ekker S C, Ungar A R, Greenstein P, von Kessler D P, Porter J A, Moon R T, Beachy P A. Curr Biol. 1995;5:944–955. doi: 10.1016/s0960-9822(95)00185-0. [DOI] [PubMed] [Google Scholar]
- 3.Kos L, Chiang C, Mahon K A. Mech Dev. 1998;70:25–34. doi: 10.1016/s0925-4773(97)00168-8. [DOI] [PubMed] [Google Scholar]
- 4.Pearse R V, II, Tabin C J. J Exp Zool. 1998;282:677–690. [PubMed] [Google Scholar]
- 5.Marti E, Takada R, Bumcrot D A, Sasaki H, McMahon A P. Development (Cambridge, UK) 1995;121:2537–2547. doi: 10.1242/dev.121.8.2537. [DOI] [PubMed] [Google Scholar]
- 6.Tabata T, Kornberg T B. Cell. 1994;76:89–102. doi: 10.1016/0092-8674(94)90175-9. [DOI] [PubMed] [Google Scholar]
- 7.Yang Y, Drossopoulou G, Chuang P T, Duprez D, Marti E, Bumcrot D, Vargesson N, Clarke J, Niswander L, McMahon A, et al. Development (Cambridge, UK) 1997;124:4393–4404. doi: 10.1242/dev.124.21.4393. [DOI] [PubMed] [Google Scholar]
- 8.Porter J A, von Kessler D P, Ekker S C, Young K E, Lee J J, Moses K, Beachy P A. Nature (London) 1995;374:363–366. doi: 10.1038/374363a0. [DOI] [PubMed] [Google Scholar]
- 9.Roelink H, Porter J A, Chiang C, Tanabe Y, Chang D T, Beachy P A, Jessell T M. Cell. 1995;81:445–455. doi: 10.1016/0092-8674(95)90397-6. [DOI] [PubMed] [Google Scholar]
- 10.Porter J A, Ekker S C, Park W J, von Kessler D P, Young K E, Chen C H, Ma Y, Woods A S, Cotter R J, Koonin E V, et al. Cell. 1996;86:21–34. doi: 10.1016/s0092-8674(00)80074-4. [DOI] [PubMed] [Google Scholar]
- 11.Porter J A, Young K E, Beachy P A. Science. 1996;274:255–259. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- 12.Cooper M K, Porter J A, Young K E, Beachy P A. Science. 1998;280:1603–1607. doi: 10.1126/science.280.5369.1603. [DOI] [PubMed] [Google Scholar]
- 13.Herz J, Willnow T E, Farese R V., Jr Nat Genet. 1997;15:123–124. doi: 10.1038/ng0297-123. [DOI] [PubMed] [Google Scholar]
- 14.Incardona J P, Gaffield W, Kapur R P, Roelink H. Development (Cambridge, UK) 1998;125:3553–3562. doi: 10.1242/dev.125.18.3553. [DOI] [PubMed] [Google Scholar]
- 15.Goldstein J L, Basu S K, Brown M S. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 16.Brown M S, Faust J R, Goldstein J L. J Biol Chem. 1978;253:1121–1128. [PubMed] [Google Scholar]
- 17.Tolleshaug H, Goldstein J L, Schneider W J, Brown M S. Cell. 1982;30:715–724. doi: 10.1016/0092-8674(82)90276-8. [DOI] [PubMed] [Google Scholar]
- 18.Simons M, Keller P, De Strooper B, Beyreuther K, Dotti C G, Simons K. Proc Natl Acad Sci USA. 1998;95:6460–6464. doi: 10.1073/pnas.95.11.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bumcrot D A, Takada R, McMahon A P. Mol Cell Biol. 1995;15:2294–2303. doi: 10.1128/mcb.15.4.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamb J E, Ray F, Ward J H, Kushner J P, Kaplan J. J Biol Chem. 1983;258:8751–8758. [PubMed] [Google Scholar]
- 21.Bizzozero O A. Methods Enzymol. 1995;250:361–379. doi: 10.1016/0076-6879(95)50085-5. [DOI] [PubMed] [Google Scholar]
- 22.Duncan J A, Gilman A G. J Biol Chem. 1998;273:15830–15837. doi: 10.1074/jbc.273.25.15830. [DOI] [PubMed] [Google Scholar]
- 23.Buttke T M, Folks T M. J Biol Chem. 1992;267:8819–8826. [PubMed] [Google Scholar]
- 24.Rothblat G H, Burns C H, Conner R L, Landrey J R. Science. 1970;169:880–882. doi: 10.1126/science.169.3948.880. [DOI] [PubMed] [Google Scholar]
- 25.Hammond C, Helenius A. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 26.Lee J J, Ekker S C, von Kessler D P, Porter J A, Sun B I, Beachy P A. Science. 1994;266:1528–1537. doi: 10.1126/science.7985023. [DOI] [PubMed] [Google Scholar]
- 27.Keller P, Simons K. J Cell Biol. 1998;140:1357–1367. doi: 10.1083/jcb.140.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bizzozero O A. Methods Enzymol. 1995;250:361–379. doi: 10.1016/0076-6879(95)50085-5. [DOI] [PubMed] [Google Scholar]