Abstract
Opposing cellular responses are typically regulated by distinct sets of genes. However, tissue transglutaminase (TGase) provides an interesting example of a single gene product that has been implicated both in affording protection against cellular insults as well as in promoting cell death. Here, we shed some light on how these conflicting activities might be manifested by demonstrating that alternative transcripts of TGase differentially affect cell viability. We show that although the full-length TGase protein affords strong protection against cell death signals, a shorter version of TGase that is truncated at the 3′ end, and thus called TGase-short (TGase-S), is cytotoxic. The apoptotic activity of TGase-S is not dependent on its transamidation activity because the mutation of a cysteine residue that is essential for catalyzing this reaction does not compromise the ability of TGase-S to induce cell death. Intriguingly, TGase-S undergoes inappropriate oligomer formation in cells before cell death, suggesting a novel mechanism for the apoptotic effects of this protein.
Keywords: aggregation, cell death, transamidation
Tissue transglutaminase (TGase) catalyzes a Ca2+-dependent transamidation reaction resulting in the formation of covalent cross-links between certain proteins or proteins and polyamines (1–3). The induction of TGase expression and enzymatic transamidation activity in cells is a highly regulated process initiated by exposure to cellular stresses, differentiation agents, or growth factors (4–9). When properly regulated, TGase-catalyzed transamidation activity plays a role in a range of physiological processes, whereas conditions that give rise to deregulated enzymatic activity have been linked to several pathologies (1–3). Enhanced TGase expression and transamidation activity are hallmarks of diseases characterized by proteinaceous aggregates, including Alzheimer's, Huntington's, and Parkinson's disease (5, 8, 10–12). Moreover, TGase has been shown to cross-link components of the protein oligomers associated with each of these neurodegenerative disorders (8, 11, 13–15).
These findings, when coupled with the fact that interfering with TGase-catalyzed transamidation activity suppresses protein aggregation and extends the survival rates of models of neurodegenerative diseases, suggest that TGase has a causative role in inducing cell death (14–16). However, the up-regulation of TGase expression and/or its catalytic activity is not detrimental to all cells. For example, TGase is overexpressed in certain types of human cancers, and the chronic TGase-catalyzed transamidation activity associated with some cancer cell lines promotes chemoresistance (4, 17–19). The ability of TGase to protect the retinoblastoma gene product from caspase-promoted degradation may help confer a survival advantage to cells (20). This may be related to recent findings that show that TGase suppresses thapsigargin-induced apoptosis of HCT116 cancer cells by cross-linking caspase 3 into a nonfunctional oligomer (21). Indeed, the apparent connections between the up-regulation and activation of TGase and the development of the malignant state have led to the idea that specific inhibitors of TGase's transamidation activity might offer a therapeutic strategy against human cancers (22).
The often contradictory cellular functions attributed to TGase are puzzling, and they raise questions regarding how its transamidation activity might account for such opposing biological outcomes as cell survival versus apoptosis. Thus, we have considered other possible mechanisms to explain these divergent functions. One possibility that could explain some of the disparities involves alternative processing of the TGase transcript. In fact, several groups have reported the identification of a novel TGase RNA transcript whose expression can be induced in cells by cytokines and is detected in the brains of Alzheimer's patients (7, 23–25). This transcript, TGase-short (TGase-S), encodes a truncated form of TGase that lacks the carboxyl-terminal (C-terminal) 138 aa. We now show that TGase-S exerts diametrically opposite effects on cell viability compared with its full-length counterpart because the latter protein confers a strong survival advantage to cells whereas TGase-S is apoptotic. Interestingly, the ability of TGase-S to induce cell death is not dependent on transamidation activity, but rather it may be the outcome of its unique capability to undergo higher-order aggregation.
Results
As a possible explanation for the distinct outcomes attributed to TGase, we compared the cellular functions of the full-length protein with the shorter splice variant, TGase-S, that is truncated from the C-terminal end (Fig. 1a). Fig. 1b shows that although the expression of both Myc-tagged TGase and TGase-S was detected in NIH 3T3 cells 48 h after transfection, the Myc-TGase protein levels were much higher. When examined at shorter time intervals after transfection, Myc-TGase-S expression was optimal at 12 h after transfection, and then it underwent a steady decline (Fig. 1b Middle). We considered whether the down-regulation of TGase-S expression was due to a selective pressure, as might be the case if TGase-S induced cell death. The morphology of cells expressing Myc-TGase-S for 48 h appeared rounded and irregular compared with cells expressing Myc-TGase (Fig. 1c Inset). Moreover, ≈65% of the cells expressing Myc-TGase-S had nuclei that were condensed or blebbed, a hallmark of cell death (Fig. 1c). The adverse effects of TGase-S on cell viability were not limited to NIH 3T3 cells, but they were also observed in SKBR3 breast cancer cells (Fig. 1d). In contrast, Myc-TGase expression protected NIH 3T3 cells from serum deprivation-induced apoptosis (Fig. 1c), consistent with previous findings (20, 26, 27).
Fig. 1.
TGase and TGase-S differently impact cell viability. (a) Schematic diagram of the functional domains of TGase (Upper) and TGase-S (Lower). The numbers refer to amino acid residues. (b) Myc-TGase or Myc-TGase-S was expressed in NIH 3T3 cells for various times and then lysed. The cell lysates were immunoblotted as indicated. (c) NIH 3T3 cells transiently expressing Myc-TGase or Myc-TGase-S were maintained in medium with or without 5% serum for 1 day and then fixed. Immunofluorescence was performed on the samples by using Myc antibody and DAPI to detect transfectants and nuclei, respectively (Inset). Cells cultured with (open bars) or without (filled bars) serum were scored for cell death as identified by nuclear condensation/blebbing and graphed. (d) SKBR3 cells expressing Myc-TGase or Myc-TGase-S for 2 days were assayed for cell death as outlined in c.
We next asked whether distinct signaling pathways regulated the expression of endogenous TGase versus TGase-S in NIH 3T3 cells. Fibroblasts were exposed to retinoic acid (RA), TNFα, or doxorubicin, and then the protein levels of the isoforms were determined. Doxorubicin induced the expression of TGase-S, and to a lesser extent, TGase, whereas RA increased only TGase expression while TNFα specifically up-regulated TGase-S expression (Fig. 2a, top two panels). Because TNFα distinctively augments the expression of the smaller TGase isoform and activates caspase 3 (Fig. 2a, third panel from top), we examined whether knocking down TGase-S by RNAi (Fig. 2b Right) affected TNFα-mediated cell death. Fig. 2c shows that ≈80% of the cells challenged with TNFα underwent cell death. However, cells transfected with TGase-RNAi-1 or 2 were less susceptible to TNFα (Fig. 2c). A similar approach was used to assess how interfering with RA-induced TGase expression influenced cell viability (Fig. 2b Left). Although treatment with RA alone did not adversely affect the cells, expression of either TGase-RNAi caused between a 2.5- and 3.5-fold increase in RA-mediated cell death over the control (Fig. 2c). Taken together, these data argue that TGase promotes, but TGase-S attenuates, cell viability.
Fig. 2.
TNFα-induced TGase-S expression promotes cell death. (a) NIH 3T3 cells were treated with 5 μM RA, 35 ng/ml TNFα, or 0.2 μM doxorubicin (Dox) for 1 or 2 days and then lysed. The cell lysates were immunoblotted as indicated. Note the presence of a larger TGase species of ≈180 kDa (TGase-S oligomer) in lysates from cells treated with TNFα or doxorubicin. (b and c) Cells transfected with TGase-RNAi-1 or -2 were treated with 5 μM RA or 35 ng/ml TNFα for 1 day and then either lysed or fixed. (b) The lysed cells were immunoblotted as indicated. (c) The fixed cells were stained with DAPI and then scored for cell death as identified by nuclear condensation/blebbing.
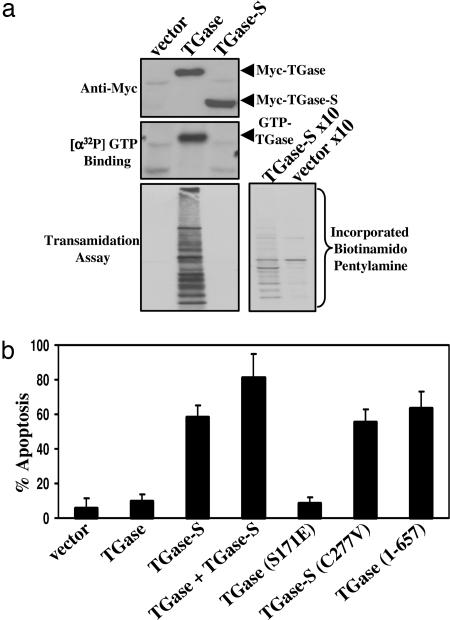
We then examined whether the transamidation or GTP-binding activity of TGase-S was responsible for its apoptotic effects. Whereas exogenously expressed Myc-TGase was enzymatically active, as read by the incorporation of 5-(biotinamido)pentylamine into proteins from cell extracts (Fig. 3a Bottom Left), Myc-TGase-S exhibited weak transamidation activity that was only detectable when using a 10-fold excess of lysate proteins (Fig. 3a Bottom Right). Likewise, consistent with previous reports (4, 26), exogenously expressed Myc-TGase showed constitutive GTP-binding activity as indicated by the incorporation of [α-32P]GTP (Fig. 3a Middle). In contrast, Myc-TGase-S failed to incorporate [α-32P]GTP. Thus, the apoptotic-promoting effects of TGase-S cannot be attributed to excessive transamidation activity or to the binding of GTP.
Fig. 3.
TGase-S-induced cell death does not require its transamidation activity. (a) Lysates from Cos7 cells expressing Myc-TGase or Myc-TGase-S were immunoblotted as indicated. The GTP-binding activities of Myc-TGase and Myc-TGase-S were determined by using an affinity-labeling assay with [α-32P]GTP. Shown is an autoradiogram of the results. The transamidation activities of the TGase isoforms were determined from the same cell lysates by assaying the incorporation of 5-(biotinamido)pentylamine into proteins. Shown are the results from an experiment where 7.5 or 75 μg (x10) of the indicated cell lysates were assayed. (b) NIH 3T3 cells expressing the indicated TGase and TGase-S constructs for 1.5 days were fixed and subjected to immunofluorescence with Myc antibody and DAPI. Transfectants were scored for cell death as identified by nuclear condensation/blebbing.
We considered whether the inability of TGase-S to bind GTP and/or its weak transamidation activity might account for its cytotoxic effects. However, these possibilities seemed unlikely, given that the expression of a point mutant of TGase, defective in both GTP-binding and transamidation activities (Myc-TGase S171E) (27), did not mirror the cell death effects caused by TGase-S (Fig. 3b). We then asked whether the adverse effects of TGase-S might be a consequence of its lacking the C-terminal portion of TGase. Expression of a TGase construct lacking 30 aa from the C-terminal end [designated TGase (1–657)] induced a cell death response comparable to that elicited by TGase-S (Fig. 3b). Furthermore, TGase (1–657), like TGase-S, was unable to bind GTP and exhibited greatly reduced transamidation activity (Fig. 4a Top) (data not shown). This emphasizes the importance of the last 30 aa of TGase in promoting its GTP-binding and transamidation activities and demonstrates that the deletion of this segment is sufficient to convert TGase into a potent death factor.
Fig. 4.
TGase-S oligomerizes in cells. Myc-tagged TGase, TGase-S, TGase (1–657), and TGase-S (C277V) were expressed in Cos7 cells and then lysed. (a) The cell lysates were immunoblotted as indicated. Note the presence of larger Myc-TGase-S and Myc-TGase (1–657) species of ≈180–240 kDa [Myc-TGase-S and TGase (1–657) Oligomers]. The GTP-binding activities of the various constructs were determined by using an affinity-labeling assay with [α-32P]GTP. Shown is an autoradiogram of the assay. (b) The indicated cell lysates were immunoblotted with TGase and actin antibodies. The larger Myc-TGase-S species detected is denoted as “TGase-S Oligomers.”
How does the C-terminal truncation of TGase yield a protein that triggers cell death? We obtained a clue when lysates from cells expressing the Myc-tagged TGase isoforms and TGase (1–657) were subjected to Western blot analysis. Fig. 4a Middle shows that in addition to the monomeric form of each of the TGase constructs, higher molecular mass forms of TGase-S and TGase (1–657) that ranged in size from ≈180 to 220 kDa were also detected. The larger-than-expected forms of Myc-tagged TGase-S and TGase (1–657) most likely reflected either the autocross-linking of these TGase molecules or their covalent cross-linking to other proteins. Moreover, the appearance of the Myc-TGase-S oligomer was not an artifact of overexpressing TGase-S, because induction of endogenous TGase-S expression with TNFα or doxorubicin resulted in the appearance of similar higher molecular mass TGase species (Fig. 2a, Top Right).
Although TGase-S displays a greatly reduced transamidation activity relative to TGase, this limited cross-linking capability might still be sufficient to give rise to the TGase-S oligomers observed by Western blot analysis. Thus, we generated a catalytically defective form of TGase-S [TGase-S (C277V)] (4, 6, 9, 20) and expressed it in cells. The Western blot in Fig. 4b shows that despite Myc-TGase-S and Myc-TGase-S (C277V) being expressed equally, TGase oligomers were only detected in lysates from cells expressing Myc-TGase-S. We then asked whether the transamidation activity of TGase-S was required for its apoptotic activity. Fig. 3b shows that cells overexpressing Myc-TGase-S (C277V) for 48 h were just as likely to undergo apoptosis as cells expressing Myc-TGase-S, indicating that the ability of TGase-S to covalently cross-link itself or other proteins is not essential for its apoptotic-promoting activity.
We still wondered whether the potential for TGase-S to undergo an autocross-linking reaction might be a clue to the mechanistic basis of its apoptotic activity, for example, by possibly influencing its cellular localization. NIH 3T3 cells, after expressing the truncated TGase isoform for 48 h, appeared small and rounded (Fig. 1c Inset), and they were not useful for determining the cellular localization of Myc-TGase-S. Thus, immunofluorescent studies were carried out on cells expressing the various TGase constructs for ≈12 h, a length of time sufficient for the induction of Myc-TGase-S expression (Fig. 1b), but preceding the onset of cell death. Fig. 5a shows the top and side views of cells expressing Myc-tagged TGase and TGase-S. Myc-TGase was uniformly expressed throughout the cytoplasm, while Myc-TGase-S aggregated within the cells. The number of aggregates observed per cell typically ranged from one to five and often displayed perinuclear localization (Fig. 5). Myc-TGase-S aggregates, however, did not colocalize with the ubiquitous Golgi protein β-COP, indicating that the unique distribution of Myc-TGase-S surrounding the nucleus was not due to its accumulation in the Golgi (Fig. 5b).
Fig. 5.
TGase-S forms large aggregates in cells. About 12 h after the transfection of NIH 3T3 cells with the indicated TGase and TGase-S constructs, the cells were fixed. (a) Immunofluorescence was performed on the samples by using Myc antibody. The top and side views of the transfectants are shown. (b) Immunofluorescence was also performed on samples by using Myc and β-COP antibodies and DAPI to detect transfectants, Golgi, and nuclei, respectively.
The analysis was expanded to determine whether the transamidation-defective form of TGase-S [Myc-TGase-S (C227V)] and the truncated form of TGase lacking the C-terminal 30 aa [Myc-TGase (1–657)] aggregated within cells. Fig. 5b indicates that expression of either of these constructs resulted in the formation of aggregates that were similar to those observed in cells expressing TGase-S. This line of evidence raises two key points. First, transamidation activity is not required for the aggregation of TGase-S in cells. Second, all forms of TGase and TGase-S that aggregate in cells also induce cell death (see Fig. 3b). Thus, inappropriate protein oligomerization may underlie the apoptotic activity of TGase-S.
To characterize further the ability of TGase-S to oligomerize in cells, extracts of cells expressing Myc-TGase or Myc-TGase-S were examined by native gel electrophoresis and gel filtration chromatography. The Myc-tagged TGase isoforms were expressed at nearly equivalent levels (Fig. 6a), and then lysates from the transfectants were resolved on a native gel. Unlike TGase, the majority of TGase-S exhibited a retarded migration such that it was detected near the wells of the gel (Fig. 6b), thus reflecting its ability to form large oligomers in cells. Fig. 6c shows the elution profiles of the TGase isoforms obtained by gel filtration chromatography of the cell extracts. As earlier reported (28), Myc-TGase eluted at ≈180 kDa, suggesting that it may exist as a dimer or associate with additional proteins in cells. In contrast, Myc-TGase-S eluted with an apparent size of ≈550 kDa. Thus, the findings from three independent approaches, namely immunofluorescence, native gel electrophoresis, and column chromatography, all support the idea that TGase-S forms oligomers when expressed in cells.
Fig. 6.
TGase and TGase-S exhibit distinct biochemical properties. Lysates of Cos7 cells expressing Myc-TGase or Myc-TGase-S were separated by SDS/PAGE (a) or native-PAGE (b) and immunoblotted as indicated. (c) The cell extracts were also loaded onto a Superdex 200 gel filtration column and eluted from the column in 1.0-ml fractions. Aliquots (50 μl) from each fraction (indicated by numbers) were immunoblotted with Myc antibody. The elution profiles of Myc-TGase-S (Upper) and Myc-TGase (Lower) are shown. Fractions 15–21 correspond to a molecular mass range from 580–500 kDa, whereas fractions 28–35 correspond to a molecular mass range from 220–140 kDa.
Discussion
TGase has been suggested to participate in a host of biological activities, including the propagation of signals that contribute to cell survival in some contexts but cell death in others (1–3). The possible connections between TGase-mediated cell survival and malignancy may be especially relevant because there have been increasing indications that inhibiting TGase's transamidation activity can enhance the actions of apoptotic factors on cancer cells (4, 17, 19, 21, 22). Unfortunately, this picture has been made confusing by reports suggesting that TGase contributes to cell death through the deregulation of its transamidation activity (9, 14–16, 29, 30). This led us to consider the possibility that some of the opposing effects on cell viability attributed to TGase might involve the existence of multiple transcripts that exhibit distinct functional properties (7, 23, 24). Although there is a good deal of precedent for different transcripts exhibiting varying degrees of a particular activity, the ability of distinct transcripts from a common gene to mediate diametrically opposing functions is rare. However, this is exactly what appears to be the case for TGase and its shorter form, TGase-S. Consistent with previous findings from our laboratory and others, expression of full-length TGase in cancer cells as well as in fibroblasts provides protection against apoptotic challenges (4, 17, 19–21, 26). In contrast, ectopic expression of TGase-S induced an apoptotic response. The initial indication for this came from our observations of an apparent selective pressure against the expression of TGase-S in fibroblasts. We then showed that TNFα selectively up-regulated TGase-S expression, whereas knocking-down TGase-S caused a partial reduction in TNF-α-induced apoptosis.
The apoptotic activity of TGase-S cannot be attributed to either of the two established activities of the full-length protein, namely the ability to bind GTP and to catalyze transamidation activity. In fact, we are unable to detect measurable GTP-binding activity for TGase-S, most likely because of the C-terminal truncation removing a portion of the guanine nucleotide-binding site (7, 31). TGase-S also exhibits only a weak transamidation activity; moreover, mutation of a cysteine residue essential for transamidation activity had no effect on its apoptotic activity.
These findings then lead to the obvious question of “how does TGase-S induce apoptosis?” The answer may be related to a rather unusual property of this isoform to exhibit higher order aggregation. This was first demonstrated through the ability of TGase-S possibly to undergo an autotransamidation reaction, resulting in the appearance of higher molecular weight forms of the protein on SDS/PAGE gels. We then found that this activity was correlated with the ability of TGase-S to form aggregates and that the aggregation of TGase-S was visualized in cells under conditions where it induced cell death.
Thus, our findings now suggest that although full-length TGase typically provides a protective effect against cellular insults and apoptotic challenges, because of its transamidation activity, the TGase-S isoform gives rise to cell death, apparently through its aberrant aggregation. Inappropriate protein oligomerization has emerged as an increasingly common mechanism for inducing cell death (11, 15). Misfolded, mutated, or posttranslationally modified proteins that acquire the ability to oligomerize indiscriminately can accumulate in cells until normal cellular processes are disrupted to the extent that cell death ensues. All of this is particularly intriguing given the suggestions that TGase-S is a possible participant in neurodegenerative disorders (23). However, this also raises a number of important questions for future study. What is the specific mechanism by which TGase-S undergoes aggregation? Can we identify specific amino acid residues that are essential for aggregation, and if so, will mutations of these sites block aggregation and consequently render TGase-S incapable of causing apoptosis? Might it ultimately be possible to identify small molecules that disrupt the aggregation and thus use this as a strategy to prevent cytotoxic events linked to various neurodegenerative disorders?
Methods
Materials.
TNFα, doxorubicin, RA, β-COP antibody, and DAPI were obtained from Sigma, and 5-(biotinamido)pentylamine was obtained from Pierce. The TGase and actin antibodies were from NeoMarkers, the Myc antibody was from Covance, and the ERK and active caspase-3 antibodies were from Cell Signaling. The stealth control-RNAi and TGase-RNAis and all cell culture reagents were from Invitrogen.
Plasmid Construction.
Full-length cDNAs encoding TGase and TGase-S (also referred to as TGase-isoform b or TGase-homologue) were cloned from HeLa cells and ligated into pcDNA3 (Invitrogen). The indicated mutations were introduced by site-directed mutagenesis using the QuikChange kit (Stratagene) or by PCR-based technologies.
Cell Culture.
NIH 3T3 and Cos7 cells were grown in Dulbecco's modified Eagle's medium containing 10% calf serum. SKBR3 cells were grown in RPMI medium containing 10% FBS. The Myc-TGase and Myc-TGase-S constructs and the TGase RNAis were introduced into cells by using Lipofectamine 2000 (Invitrogen). Cells were lysed with cell lysis buffer (25 mM Tris/100 mM NaCl/1% Triton X-100/1 mM EDTA/1 mM DTT/1 mM NaVO4/1 μg/ml each aprotinin and leupeptin).
Western Blot Analysis.
Cell lysates were subjected to SDS/PAGE, and then the proteins were transferred to PDVF. The filters were incubated with the various primary antibodies diluted in TBST (20 mM Tris/137 mM NaCl, pH 7.4/0.02% Tween 20). The primary antibodies were then detected with horseradish peroxidase-conjugated secondary antibodies followed by exposure to ECL.
Transamidation Assay and Photoaffinity Labeling of TGase.
Immunofluorescence and Cell Death Assay.
Cells transfected with the various expression plasmids and/or exposed to various culturing conditions were fixed with 5% formaldehyde, permeabilized with PBS containing 0.1% Triton X-100, and probed with the indicated primary antibodies. Primary antibodies were detected with either Oregon green 488- or rhodamine red-conjugated secondary antibodies (Molecular probes), and DAPI was used to stain nuclei. The cells were visualized by fluorescence microscopy. Cells undergoing cell death were identified by nuclear condensation/blebbing.
Gel Filtration.
Cos7 cells expressing Myc-TGase or Myc-TGase-S were lysed by using lysis buffer devoid of Triton X-100. Myc-TGase and Myc-TGase-S were precipitated from the cell extracts by using 80% (NH4)2SO4, resuspended in buffer A (20 mM Tris, pH 8.0/150 mM NaCl/2 mM DTT) and then dialyzed in the same buffer. The samples were then loaded onto a fast protein liquid chromatography Superdex-200 Highload 16/60 column, and 1-ml fractions were collected. Aliquots (50 μl) from each fraction were subjected to SDS/PAGE, and the proteins were transferred to PDVF. The filters from the native-PAGE experiment and the gel filtration fractions were probed with anti-Myc antibody.
Acknowledgments
This work was supported by National Institutes of Health Grant GM61762.
Abbreviations
- RA
retinoic acid
- TGase
tissue transglutaminase
- TGase-S
TGase-short.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Griffin M, Casadio R, Bergamini CM. Biochem J. 2002;368:377–396. doi: 10.1042/BJ20021234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lorand L, Graham RM. Nat Rev Mol Cell Biol. 2003;4:140–156. doi: 10.1038/nrm1014. [DOI] [PubMed] [Google Scholar]
- 3.Fesus L, Szondy Z. FEBS Lett. 2005;579:3297–3302. doi: 10.1016/j.febslet.2005.03.063. [DOI] [PubMed] [Google Scholar]
- 4.Antonyak MA, Miller AM, Jansen JM, Boehm JE, Balkman CE, Wakshlag JJ, Page RL, Cerione RA. J Biol Chem. 2004;279:41461–41467. doi: 10.1074/jbc.M404976200. [DOI] [PubMed] [Google Scholar]
- 5.Lesort M, Chun W, Johnson GV, Ferrante RJ. J Neurochem. 1999;73:2018–2027. [PubMed] [Google Scholar]
- 6.Tucholski J, Lesort M, Johnson GV. Neuroscience. 2001;102:481–491. doi: 10.1016/s0306-4522(00)00482-6. [DOI] [PubMed] [Google Scholar]
- 7.Monsonego A, Shani Y, Friedmann I, Paas Y, Eizenberg O, Schwartz M. J Biol Chem. 1997;272:3724–3732. doi: 10.1074/jbc.272.6.3724. [DOI] [PubMed] [Google Scholar]
- 8.Karpuj MV, Garren H, Slunt H, Price DL, Gusella J, Becher MW, Steinman L. Proc Natl Acad Sci USA. 1999;96:7388–7393. doi: 10.1073/pnas.96.13.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tucholski J, Johnson GV. J Neurochem. 2002;81:780–791. doi: 10.1046/j.1471-4159.2002.00859.x. [DOI] [PubMed] [Google Scholar]
- 10.Kim SY, Grant P, Lee JH, Pant HC, Steinert PM. J Biol Chem. 1999;274:30715–30721. doi: 10.1074/jbc.274.43.30715. [DOI] [PubMed] [Google Scholar]
- 11.Junn E, Ronchetti RD, Quezado MM, Kim SY, Mouradian MM. Proc Natl Acad Sci USA. 2003;100:2047–2052. doi: 10.1073/pnas.0438021100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson GV, Cox TM, Lockhart JP, Zinnerman MD, Miller ML, Powers RE. Brain Res. 1997;751:323–329. doi: 10.1016/s0006-8993(96)01431-x. [DOI] [PubMed] [Google Scholar]
- 13.Kahlem P, Green H, Djian P. Mol Cell. 1998;1:595–601. doi: 10.1016/s1097-2765(00)80059-3. [DOI] [PubMed] [Google Scholar]
- 14.Mastroberardino PG, Iannicola C, Nardacci R, Bernassola F, De Laurenzi V, Melino G, Moreno S, Pavone F, Oliverio S, Fesus L, Piacentini M. Cell Death Differ. 2002;9:873–880. doi: 10.1038/sj.cdd.4401093. [DOI] [PubMed] [Google Scholar]
- 15.Dedeoglu A, Kubilus JK, Jeitner TM, Matson SA, Bogdanov M, Kowall NW, Matson WR, Cooper AJ, Ratan RR, Beal MF, et al. J Neurosci. 2002;22:8942–8950. doi: 10.1523/JNEUROSCI.22-20-08942.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karpuj MV, Becher MW, Springer JE, Chabas D, Youssef S, Pedotti R, Mitchell D, Steinman L. Nat Med. 2002;8:143–149. doi: 10.1038/nm0202-143. [DOI] [PubMed] [Google Scholar]
- 17.Herman JF, Mangala LS, Mehta K. Oncogene. 2006;25:3049–3058. doi: 10.1038/sj.onc.1209324. [DOI] [PubMed] [Google Scholar]
- 18.Zhang R, Tremblay TL, McDermid A, Thibault P, Stanimirovic D. Glia. 2003;42:194–208. doi: 10.1002/glia.10222. [DOI] [PubMed] [Google Scholar]
- 19.Yuan L, Choi K, Khosla C, Zheng X, Higashikubo R, Chicoine MR, Rich KM. Mol Cancer Ther. 2005;4:1293–1302. doi: 10.1158/1535-7163.MCT-04-0328. [DOI] [PubMed] [Google Scholar]
- 20.Boehm JE, Singh U, Combs C, Antonyak MA, Cerione RA. J Biol Chem. 2002;277:20127–20130. doi: 10.1074/jbc.C200147200. [DOI] [PubMed] [Google Scholar]
- 21.Yamaguchi H, Wang HG. Mol Cell Biol. 2006;26:569–579. doi: 10.1128/MCB.26.2.569-579.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi K, Siegel M, Piper JL, Yuan L, Cho E, Strnad P, Omary B, Rich KM, Khosla C. Chem Biol. 2005;12:469–475. doi: 10.1016/j.chembiol.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Citron BA, Suo Z, SantaCruz K, Davies PJ, Qin F, Festoff BW. Neurochem Int. 2002;40:69–78. doi: 10.1016/s0197-0186(01)00062-6. [DOI] [PubMed] [Google Scholar]
- 24.Festoff BW, SantaCruz K, Arnold PM, Sebastian CT, Davies PJ, Citron BA. J Neurochem. 2002;81:708–718. doi: 10.1046/j.1471-4159.2002.00850.x. [DOI] [PubMed] [Google Scholar]
- 25.Fraij BM, Birckbichler PJ, Patterson MK, Jr, Lee KN, Gonzales RA. J Biol Chem. 1992;267:22616–22623. [PubMed] [Google Scholar]
- 26.Antonyak MA, McNeill CJ, Wakshlag JJ, Boehm JE, Cerione RA. J Biol Chem. 2003;278:15859–15866. doi: 10.1074/jbc.M300037200. [DOI] [PubMed] [Google Scholar]
- 27.Antonyak MA, Singh US, Lee DA, Boehm JE, Combs C, Zgola MM, Page RL, Cerione RA. J Biol Chem. 2001;276:33582–33587. doi: 10.1074/jbc.M105318200. [DOI] [PubMed] [Google Scholar]
- 28.Singh US, Cerione RA. J Biol Chem. 1996;271:27292–27298. doi: 10.1074/jbc.271.44.27292. [DOI] [PubMed] [Google Scholar]
- 29.Oliverio S, Amendola A, Rodolfo C, Spinedi A, Piacentini M. J Biol Chem. 1999;274:34123–34128. doi: 10.1074/jbc.274.48.34123. [DOI] [PubMed] [Google Scholar]
- 30.Melino G, Annicchiarico-Petruzzelli M, Piredda L, Candi E, Gentile V, Davies PJ, Piacentini M. Mole Cell Biol. 1994;14:6584–6596. doi: 10.1128/mcb.14.10.6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Begg GE, Holman SR, Stokes PH, Matthews JM, Graham RM, Iismaa SE. J Biol Chem. 2006;281:12603–12609. doi: 10.1074/jbc.M600146200. [DOI] [PubMed] [Google Scholar]