Abstract
Background
A key event in the origin of life on this planet has been formation of self-replicating RNA-type molecules, which were complex enough to undergo a Darwinian-type evolution (origin of the "RNA world"). However, so far there has been no explanation of how the first RNA-like biopolymers could originate and survive on the primordial Earth.
Results
As condensation of sugar phosphates and nitrogenous bases is thermodynamically unfavorable, these compounds, if ever formed, should have undergone rapid hydrolysis. Thus, formation of oligonucleotide-like structures could have happened only if and when these structures had some selective advantage over simpler compounds. It is well known that nitrogenous bases are powerful quenchers of UV quanta and effectively protect the pentose-phosphate backbones of RNA and DNA from UV cleavage. To check if such a protection could play a role in abiogenic evolution on the primordial Earth (in the absence of the UV-protecting ozone layer), we simulated, by using Monte Carlo approach, the formation of the first oligonucleotides under continuous UV illumination. The simulations confirmed that UV irradiation could have worked as a selective factor leading to a relative enrichment of the system in longer sugar-phosphate polymers carrying nitrogenous bases as UV-protectors. Partial funneling of the UV energy into the condensation reactions could provide a further boost for the oligomerization.
Conclusion
These results suggest that accumulation of the first polynucleotides could be explained by their abiogenic selection as the most UV-resistant biopolymers.
Background
In modern concepts of the origin of life, there is an glaring gap between the abiogenic formation of the first building blocks and the origin of the "RNA world" i.e. of the first RNA-like polynucleotides that could undergo a Darwinian-type evolution [1-5]. Indeed, there is a wealth of experimental evidence for the abiogenic formation of amino acids, nitrogenous bases and carbohydrates from inorganic compounds like cyanide, thiocyanate, and carbon monoxide under reducing and/or neutral conditions [6-9] (reviewed in ref. [10]). On the other hand, the documented catalytic activity of RNA molecules [11,12] allows to suggest that primordial ribonucleotides could have initially evolved on their own, without assistance from proteins [2-4,13]. What is missing is a physically plausible mechanism for the thermodynamically unfeasible event of formation and accumulation of long oligonucleotide-like polymers.
This problem can be focused even further. Aluminosilicate clays have been shown to catalyze the formation of oligonucleotides of up to 50 units long, when supplied by preformed and pre-activated mononucleotides under optimized laboratory conditions [14-16]. However, no oligonucleotide formation from pentose phosphates and nitrogenous bases has been reported so far under the supposedly primordial conditions where the formation of amino acids, nitrogenous bases and carbohydrates took place. Furthermore, the current understanding implies that the environmental conditions on the primeval Earth were unfavorable for the survival of oligonucleotide-like polymers. A particularly important factor is that, due to the absence of the ozone layer, the UV flux at the Earth surface must have been approximately 100 times larger than it is now [17,18], causing deterioration of most organic molecules. The existing theories consider the high UV level as a major obstacle and offer several different strategies for hiding the first life forms from it (see e.g. ref. [10,19,20]). Here we invoke an alternative possibility, i.e. that the UV irradiation played a positive role in the origin of life by serving as a principal selective factor in the formation of pre-biological structures. Moreover, the influx of energy into the system in the form of the UV irradiation could be seen as the driving force required for the gradual complication of the system [1]. These considerations prompted us to analyze the possible effects of the UV irradiation on oligonucleotide formation in primordial conditions.
Results
Nitrogen bases as efficient UV-quenchers
In general terms, UV irradiation of first RNA-like polymers could be damaging for their nitrogenous bases, their pentose-phosphate backbone and for the bonds between the bases and the backbone. It is known that the ether bonds in monomeric sugar phosphates are much more susceptible to UV damage than monomeric nitrogenous bases: e.g. the quantum yields of UV damage by 254 nm irradiation for sugar phosphates and monomeric nitrogenous bases are approx. 10-2 and 10-4, respectively (see ref. [21] for a comprehensive review). However, numerous studies of modern nucleic acids revealed an interesting paradox: nitrogenous bases, both purines and pyrimidines, of DNA and RNA are much more sensitive to the UV illumination than the pentose-phosphate backbone. It appears therefore that nitrogenous bases protect the pentose-phosphate backbone from the UV damage. Indeed, nitrogenous bases get rid of the absorbed UV energy extremely fast. Their fluorescence life times at room temperature are on the order of 10-12 sec [21]. This means that within 1 picosecond the energy of trapped UV quanta is dissipated, predominantly into heat. As a result, the probability of a transition into a potentially photochemically active, long-living triplet state is quite low, on the order of 10-3 at room temperature [21]. For comparison, the fluorescence life time of tryptophan, which has a comparably complex structure, is more than 1000-fold longer, on the order of 5 ns. Correspondingly, the quantum yield of the singlet → triplet transition is higher, about 0.2 at room temperature [32]. The extremely efficient deactivation of the UV quanta by nitrogenous bases allows them to protect the compounds to which they are attached from UV-induced breakage. It has been demonstrated that the photo-cleavage of ortophosphate from the sugar moiety of AMP in response to irradiation by a mercury lamp with an emission at 254 nm proceeded (with a quantum yield of 5× 10-5 mol/Einstein) about 10 times slower than from glycerol 2-phosphate [22], although AMP molecules, because of their higher molar extinction at 254 nm, were hit by 103–104 times more quanta per unit of time as compared to glycerol 2-phosphate molecules. These data were explained by a prompt one-way spillover of the excitation energy adsorbed by sugar-phosphate moiety of AMP to the nitrogenous base [22]. The latter apparently served as an excellent sink for the UV energy because of the lower energy of the first singlet level as compared to sugar phosphates. In RNA from tobacco mosaic virus, the quantum yield for the breakage of the sugar-phosphate bonds by 254 nm UV light has been shown to be 3.5 × 10-6 mol/Einstein [23], which corresponds to a UV-protection factor of 70 as compared to "bare" sugar phosphates. In human skin DNA, the quantum yield for a single strand break by 254 nm UV was in the order of 10-8 [21,24], indicating an even higher backbone protection factor of about 2000. It is noteworthy that the quantum yield of photo-damage to nitrogenous bases proper in RNA and DNA is on the order of 10-4 mol/Einstein [21], i.e. it is 103–104 times higher than that of the backbone breakage. Apparently, the backbones of RNA and DNA can be rescued due to the partial "victimization" of nitrogenous bases. Further factors, which might contribute to the backbone protection, are the effective shielding of the backbone from the UV-light by nitrogenous bases, the excitonic coupling between the latter, and the elevated capacity of polymeric compounds to dissipate the excessive thermal energy without undergoing a mechanical damage.
Model of nucleotide polymerization under the conditions of UV illumination
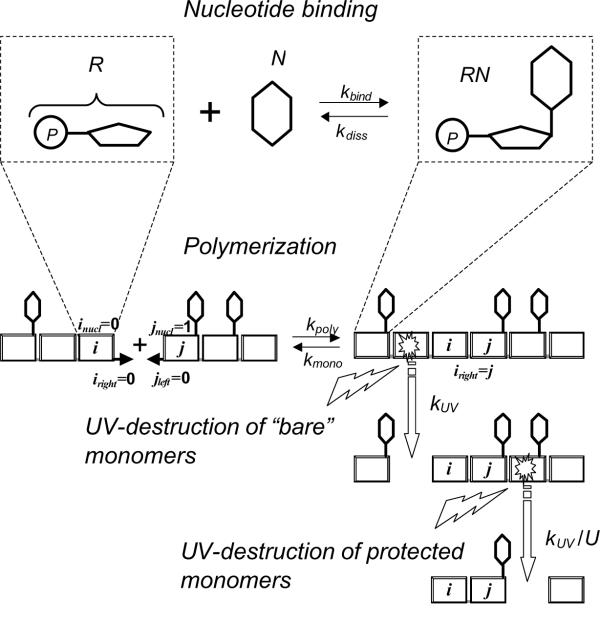
To explore the significance of such a UV protection mechanism for the evolution of primordial polymers, we modeled the polymerization of sugar-phosphate monomers (R) in the presence of nitrogenous bases (N) under the conditions of UV illumination. The exact nature of the sugar moiety was of secondary importance for the model because of approximately similar susceptibility of various sugar-phosphates to UV cleavage [21]. The set of reactions considered is schematically depicted in Figure 1. As long as a UV quantum can split a polynucleotide chain at any point, we had to use the Monte-Carlo approach. Because the system was open and the chains were only transient objects, the model topology was described by three pointers assigned to individual monomers as shown in Figure 1. Namely, the polymerization state of monomer i was characterised by two pointers ileft and iright defined as follows: if the left bond of monomer i was unsaturated, we put ileft = 0; if the monomer i was left-bound to monomer j, then ileft = j; the same rules were used for the right pointer iright. The chemical state of monomer i was characterized by an additional variable inucl that took values "1" or "0" when the given monomer was, respectively, bound or not bound to a nitrogenous base.
Figure 1.
Schematic representation of the modeled reactions
The reaction dynamics was considered in discrete time steps. The interval between steps Δt was chosen appropriately small so that differential equations of chemical kinetics could be replaced by their discrete analogues. At each step, sugar-phosphate monomers were added to the reaction volume ΔV in the quantity to keep their total concentration constant. The value of ΔV was chosen so that the system contained on average about 2000 monomers.
At each time step the following reactions were considered:
Each monomer i with unsaturated left (right) bond (ileft(right) = 0) could join a polymer chain reacting with monomer j having unsaturated right (left) bond (jright(left) = 0) under a condition that monomer j does not belong to the same chain as monomer i. The probability of such reaction was calculated as Ppoly(i, j) = kpoly·Δt·ΔV-1. Each monomer i with saturated left (right) bond (ileft(right) ≠ 0) could re-dissociate with the probability Pmono = kmono·Δt.
Because the UV stability of nitrogenous bases is by factor of 102 higher than that of not protected, "bare" sugar-phosphates (see above), the former were assumed to be present in excess. Then each R monomer could bind nitrogenous base with the probability Pbind = kbind·Δt and each RN nucleotide could dissociate with the probability Pdiss = kdiss·Δt independently of the polymerization state. The ratio of the rate constants kbind and kdiss reflects the thermodynamic equilibrium of the nucleotide binding reaction: kbind/kdiss = 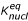 = exp(-ΔGnucl/kT).
= exp(-ΔGnucl/kT).
The probability of UV-decomposition for "bare" sugar-phosphate monomers was DUV = kUV·Δt independently of whether they were part of a polymer chain or not. A nitrogenous base protected the sugar-phosphate to which it was bound from UV degradation by a factor U so that the probability of its decomposition was DUV = U-1·kUV·Δt.
Monte-Carlo simulations
On simulation, we assumed, for simplicity, that each monomer is hit by a UV quantum once per second. On picking the values of polymerization rate constant kpoly, we assumed, in line with experimental observations [14-16], that this reaction was slow. The experimental observation of the formation of oligonucleotides of up to 50 units on clays [16-18] implies an equilibrium constant of an inorganic template-guided polymerization 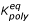 = kpoly·[R] / kmono > 1. In contrast, nitrogenous base binding was assumed to be thermodynamically unfavorable, with a respective equilibrium constant
= kpoly·[R] / kmono > 1. In contrast, nitrogenous base binding was assumed to be thermodynamically unfavorable, with a respective equilibrium constant 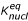 = kbind / kdiss << 1. The values of the UV-decay rate constant kUV and of the UV-protection factor U were varied around their experimentally established values of 10-1 – 10-3 and 10 – 103, respectively (see refs above).
= kbind / kdiss << 1. The values of the UV-decay rate constant kUV and of the UV-protection factor U were varied around their experimentally established values of 10-1 – 10-3 and 10 – 103, respectively (see refs above).
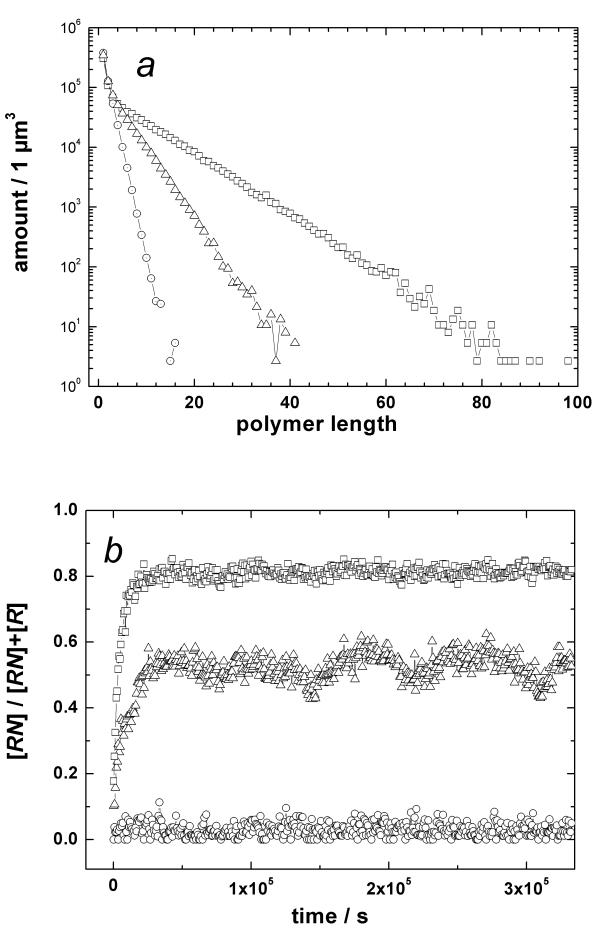
As long as parameters were varied in the indicated ranges, the simulation behavior of the system was qualitatively stable, with a typical example depicted in Figure 2. In the absence of UV protection (U = 1), the polymerization yield was marginal (Fig. 2a, circles) and the extent of nitrogenous base incorporation in the polymers was close to zero (Fig. 2b, circles). With the UV protection "switched on" (U > 1), the length of formed polymers increased dramatically (Fig. 2a, triangles). Even more importantly, under these conditions the fraction of nitrogenous-base-carrying sugar-phosphates increased significantly (up to ~0.5 in the case presented in Fig. 2b by triangles). Thus, in our simulation, the UV selection caused a strong relative enhancement in oligonucleotide-like polymers even under conditions where the binding of nitrogenous bases to sugar-phosphate moieties was thermodynamically unfavorable.
Figure 2.
Monte Carlo simulation of a sugar-phosphate polymerization reaction in the presence of nitrogenous bases and under UV-illumination Sugar-phosphate polymerization reaction in the presence of nitrogenous bases and under UV-illumination was simulated using the following parameter set: The concentration of monomers in the reaction volume was kept on a constant level of 10-3 M (comparable with their concentration in the cell). The second-order rate constant of polymerization kpoly was 3 M-1 s-1 and the first-order rate constant of re-dissociation kmono was 10-4 s-1 that corresponded to the equilibrium constant 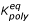 = 30. The rate constants of nucleotide binding (kbind) and dissociation (kdiss) were 3 × 10-8 s-1 and 10-6 s-1, respectively (
= 30. The rate constants of nucleotide binding (kbind) and dissociation (kdiss) were 3 × 10-8 s-1 and 10-6 s-1, respectively (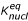 = 3 × 10-2). Under the UV illumination, monomers decomposed with the rate constant of 3·10-3 s-1 irrespectively of their position in the chain. For simplicity, the UV protection factor U of 30 was used both for monomers and oligomers. The partial funneling of UV energy was assumed to increase the kbind value from 3 × 10-8s-1 up to 1.2 × 10-7 s-1. a, Polymer length distribution at equilibrium. b, Fraction of monomers protected by nitrogenous bases as a function of time.
= 3 × 10-2). Under the UV illumination, monomers decomposed with the rate constant of 3·10-3 s-1 irrespectively of their position in the chain. For simplicity, the UV protection factor U of 30 was used both for monomers and oligomers. The partial funneling of UV energy was assumed to increase the kbind value from 3 × 10-8s-1 up to 1.2 × 10-7 s-1. a, Polymer length distribution at equilibrium. b, Fraction of monomers protected by nitrogenous bases as a function of time.
We also simulated the effect of partial funneling of UV energy into the condensation reactions. In Figure 2, we depict by squares a case where we accounted for a possibility of a productive binding of a nitrogenous base N by a photo-generated radical R•. The quantum yield of such a reaction was taken as small as 9 × 10-8. The resulting boost for the formation of oligonucleotides was remarkable concerning the length of the formed polymer chains (Fig. 2a, squares), the nitrogenous base incorporation rate, and the extent of the nitrogenous bases incorporation into oligomers (Fig. 2b, squares), so that polymers were built predominantly from nucleotides. (We neglected here the possibility that nucleotides could arise also from an interaction of light-generated nitrogenous base radicals N• with sugar-phosphates R. Because the extinction of nitrogenous bases in UV is by orders of magnitude higher than that of sugar phosphates, this possibility is more than real). The experimentally measured quantum yield (probability) of the compatible photo-condensation reaction was actually 103 times higher than we assumed in our simulation: the quantum yield of the 254 nm UV-driven AMP formation from adenine, ribose and ethyl metaphosphate reached 10-4 mol/Einstein after one hour of illumination [7]. The inclusion of the photo-condensation quantum yield values compatible to those measured in ref. [7] into the parameter set used for simulation in Fig. 2 led to an overload because of unmanageable increase in the polymer chain length. It is hard to believe that evolution has not used the opportunity to drive the primordial condensation by energy of light, with nitrogenous bases working as light-absorbing antenna.
Discussion
Thus, the results of our Monte-Carlo simulation indicate that a mechanism of natural selection, similar to the one that has driven the subsequent biological evolution, could have been responsible for the primordial polymerization. It seems quite unlikely that the extremely effective UV-quenching by all five major nucleobases is just incidental. Accordingly, one can assume that these bases had been selected to perform the UV-protecting function before they became involved in the maintenance and transfer of genetic information. This assumption provides a physically plausible rationale for the primordial enrichment in oligonucleotide-like compounds and also sheds new light on the earliest steps of evolution.
It is worth noting the following points:
(i) In the UV-illuminated primordial world the probability of a UV-breakage was more than real for any compound. Correspondingly, those that succeeded to bind (trap) a UV-quencher got a selective advantage. In the case of nitrogenous bases, the photo-generated radicals of the latter could even provide the energy needed to bind the UV-protector.
(ii) The oligomerization, most probably inorganic template-guided (see ref. [10] for a comprehensive consideration of this point), further increased the degree of the UV-protection because then each UV-trap could protect several neighbouring backbone bonds. The minerals considered as potential primordial templates (see ref. [10] for their survey) are characterized by meshes of positively charged metal ions, so that a prospective oligomer unit required a negatively charged template-binding group (phosphate was the natural choice [10]) and a spacer region needed to fit into the lattice of positively charges at the template. The pentose groups are apt spacers as long as aluminosilicate clays indeed catalyse the formation of long oligonucleotides [14-16].
(iii) In this "sugar-phosphate-centric" world the nitrogenous bases served just as protecting units, so that the UV-quenching capacity but not their exact chemical nature was important. Accordingly, these service units were replaceable and variable. Exactly this variability could have paved the way to the variability of the future genomes.
(iv) In general, the increase in the fraction of double-stranded regions elevates the resistance of RNA backbone to UV-damage [25], as well as to hydrolysis [26]. Correspondingly, an occasional formation of double-stranded segments in primordial RNA-like polymers elevated, as it has been already discussed [26], the probability of their survival. Hence the survival, apparently, depended on the ability of chosen UV-protectors to form pairs.
(v) Replication, most probably also initially template-guided [10], could have evolved as a process that just selected for more photostable biopolymers. Indeed, the first replication event, i.e. formation of a polynucleotide strand using another one as a template, could not be driven by the reproduction needs (the chicken and egg paradox). Replication could be, however, promoted by the simple fact that the UV-resistance of two interacting polynucleotide strands was higher than that of single strands.
The suggested mechanism turns the high UV levels on primordial Earth from a perceived obstacle to the origin of life (see e.g. ref. [19]) into the selective factor that, in fact, might drive the whole process. Indeed, biochemical condensation reactions proceed with release of water, so that the presence of latter favors hydrolysis of biological polymers. Because of this feature, Bernal [27] and many researchers after him (as reviewed in ref. [10]) advanced the view that life has begun in tidal regions, so that condensation of primordial monomers proceeded under "fluctuating" conditions where the wet periods, enabling the exchange of reagents, alternated with dry ones, favoring the condensation reactions. The awareness of the potential danger of the UV damage, however, prompted other scientists to invoke a UV-protecting water layer (see e.g. ref. [19]), which apparently would impede the condensation reactions. More recently, several authors even moved the point of the life origin to the bottom of the ocean, where the reducing power of minerals and/or of hydrothermal vents was considered to be the energy source for the first condensation events [28,29]. It remained unexplained, though, how inorganic reductants could drive primordial condensation reactions in water in the absence of enzymes (see the discussion in refs. [30,31]).
In a sense, the absence of a consensus on a plausible mechanism for the origin and accumulation of the first RNA-like molecules has significantly hurt the development in the whole field and stimulated proliferation of the Panspermia hypothesis, not to mention various kinds of creationist ideas. It appears that our consideration of the UV irradiation as a positive, selective factor in primordial evolution may suggest a way out of the dead end. This view allows to place the cradle of life onto the sun-illuminated (semi)dry surface of the ancient Earth, as originally considered by Bernal [27]. Indeed, no other known energy source could compete with the UV component of the solar irradiation either in ability to serve simultaneously as both selective and driving force, or in continuity, strength, and access to the whole surface of Earth.
We believe that the ideas discussed above could eventually be tested experimentally. On one hand, nucleotides could be formed from simpler compounds under conditions of UV-irradiation [7,8]. On the other hand, the ability of aluminosilicate clays to catalyze their polymerization was demonstrated [14-16]. Thus it seems very promising to try obtaining oligonucleotides from simpler compounds in a reactor system, which (i) uses UV-irradiation as a selective force and an energy source and (ii) contains aluminosilicate clays as a polymerization template.
Conclusions
In conclusion, we believe that the proposed mechanism for selective accumulation of primordial polynucleotide-like structures, coupled with the physical plausibility of their subsequent Darwinian evolution, compellingly demonstrated earlier [1-5], offers a coherent picture of a selection-driven emergence of increasingly complex biological molecules.
Methods
Modeling the nucleotide polymerization under the conditions of UV illumination
The complete simulation code in MATLAB5 (The Mathworks Inc, MA, USA) is available on request.
Authors' contributions
AYM came up with the main idea, initiated the study and drafted the manuscript. DAC created the model, programmed the simulation code and carried out the Monte-Carlo simulations. MYG helped formulating the model and participated in the discussion of the results and writing of the paper. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank Prof. Wolfgang Junge and Prof. Vladimir Skulachev for stimulating interest to this work and Dr. Eugene Koonin for helpful comments. This work has been supported in part by the Alexander von Humboldt Foundation and by grants from the Deutsche Forschungsgemeinschaft (436-RUS-113/210, SFB-436-P15) and INTAS (2001-0736). We greatly appreciate the peer review and the speed and convenience of the whole process at BMC Evolutionary Biology.
Contributor Information
Armen Y Mulkidjanian, Email: mulkidjanian@biologie.uni-osnabrueck.de.
Dmitry A Cherepanov, Email: cherepanov@biologie.uni-osnabrueck.de.
Michael Y Galperin, Email: galperin@ncbi.nlm.nih.gov.
References
- Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58:465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- Gilbert W. The RNA world. Nature. 1986;319:618. [Google Scholar]
- Orgel LE. RNA catalysis and the origins of life. J Theor Biol. 1986;123:127–149. doi: 10.1016/s0022-5193(86)80149-7. [DOI] [PubMed] [Google Scholar]
- Joyce GF. The antiquity of RNA-based evolution. Nature. 2002;418:214–221. doi: 10.1038/418214a. [DOI] [PubMed] [Google Scholar]
- Szabo P, Scheuring I, Czaran T, Szathmary E. In silico simulations reveal that replicators with limited dispersal evolve towards higher efficiency and fidelity. Nature. 2002;420:340–343. doi: 10.1038/nature01187. [DOI] [PubMed] [Google Scholar]
- Miller SL. A production of amino acids under possible primitive Earth conditions. Science. 1953;117:529. doi: 10.1126/science.117.3046.528. [DOI] [PubMed] [Google Scholar]
- Ponnamperuma C, Sagan C, Mariner R. Synthesis of adenosine triphosphate under possible primitive earth conditions. Nature. 1963;199:222–226. doi: 10.1038/199222a0. [DOI] [PubMed] [Google Scholar]
- Ponnamperuma C. Primordial organic chemistry and the origin of life. Q Rev Biophys. 1971;4:77–106. doi: 10.1017/s0033583500000603. [DOI] [PubMed] [Google Scholar]
- Lazcano A, Miller SL. The origin and early evolution of life: prebiotic chemistry, the pre-RNA world, and time. Cell. 1996;85:793–798. doi: 10.1016/s0092-8674(00)81263-5. [DOI] [PubMed] [Google Scholar]
- Lahav N. Biogenesis: Theories of Life Origin. New York: Oxford University Press; 1999. [Google Scholar]
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Cech TR. The efficiency and versatility of catalytic RNA: implications for an RNA world. Gene. 1993;135:33–36. doi: 10.1016/0378-1119(93)90046-6. [DOI] [PubMed] [Google Scholar]
- Yarus M. Boundaries for an RNA world. Curr Opin Chem Biol. 1999;3:260–267. doi: 10.1016/S1367-5931(99)80041-6. [DOI] [PubMed] [Google Scholar]
- Ferris JP, Hill AR, Jr, Liu R, Orgel LE. Synthesis of long prebiotic oligomers on mineral surfaces. Nature. 1996;381:59–61. doi: 10.1038/381059a0. [DOI] [PubMed] [Google Scholar]
- Ertem G, Ferris JP. Formation of RNA oligomers on montmorillonite: site of catalysis. Orig Life Evol Biosph. 1998;28:485–499. doi: 10.1023/A:1006510228943. [DOI] [PubMed] [Google Scholar]
- Ferris JP. Montmorillonite catalysis of 30–50 mer oligonucleotides: Laboratory demonstration of potential steps in the origin of the RNA world. Orig Life Evol Biosph. 2002;32:311–332. doi: 10.1023/A:1020543312109. [DOI] [PubMed] [Google Scholar]
- Garcia-Pichel F. Solar ultraviolet and the evolutionary history of cyanobacteria. Orig Life Evol Biosph. 1998;28:321–347. doi: 10.1023/A:1006545303412. [DOI] [PubMed] [Google Scholar]
- Cockell CS, Horneck G. The history of the UV radiation climate of the earth–theoretical and space-based observations. Photochem Photobiol. 2001;73:447–451. doi: 10.1562/0031-8655(2001)073<0447:thotur>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Sagan C. Ultraviolet selection pressure on the earliest organisms. J Theor Biol. 1973;39:195–200. doi: 10.1016/0022-5193(73)90216-6. [DOI] [PubMed] [Google Scholar]
- Cleaves HJ, Miller SL. Oceanic protection of prebiotic organic compounds from UV radiation. Proc Natl Acad Sci USA. 1998;95:7260–7263. doi: 10.1073/pnas.95.13.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet J, Vigni P. The photochemistry of nucleic acids. In: Morrison H, editor. In Bioorganic Photochemistry: Photochemistry and the Nucleic Acids. New York: John Wiley & Sons; 1990. pp. 1–273. [Google Scholar]
- Goossen JTH, Kloosterboer JG. Photolysis and hydrolisis of adenosine 5'-phosphates. Photochem Photobiol. 1978;27:703–708. [Google Scholar]
- Cochran DR, Buzzell A, Lauffer MA. The effect of ultraviolet irradiation on nucleic acids isolated from tobacco mosaic virus. Biochim Biophys Acta. 1962;55:755–767. doi: 10.1016/0006-3002(62)90854-5. [DOI] [PubMed] [Google Scholar]
- Netrawali MS, Cerutti PA. Increased near-ultraviolet induced DNA fragmentation in xeroderma pigmentosum variants. Biochem Biophys Res Commun. 1979;87:802–810. doi: 10.1016/0006-291x(79)92029-1. [DOI] [PubMed] [Google Scholar]
- Jagger J. Ultraviolet inactivation of biological systems. In: Wang SY, editor. In Photochemistry and Photobiology of Nucleic Acids. II Biology. New York: Academic Press; 1976. pp. 147–186. [Google Scholar]
- Eigen M, Winkler R. In Mannheimer Forum 73/74. Mannheim: Boehringer Mannheim GmbH; 1974. Ludus Vitalis; pp. 53–139. [Google Scholar]
- Bernal JD. The Physical Basis of Life. London: Routledge and Kegan Paul; 1951. [Google Scholar]
- Wachtershauser G. Before enzymes and templates: theory of surface metabolism. Microbiol Rev. 1988;52:452–484. doi: 10.1128/mr.52.4.452-484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Russell MJ. On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos Trans R Soc Lond B Biol Sci. 2003;358:59–85. doi: 10.1098/rstb.2002.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Duve C, Miller SL. Two-dimensional life? Proc Natl Acad Sci USA. 1991;88:10014–10017. doi: 10.1073/pnas.88.22.10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel LE. Self-organizing biochemical cycles. Proc Natl Acad Sci USA. 2000;97:12503–12507. doi: 10.1073/pnas.220406697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensasson RV, Land EJ, Truscott TG. Flash Photolysis and Pulse Radiolysis: Contributions to the Chemistry of Biology and Medicine. Oxford: Pergamon Press; 1983. [Google Scholar]