Abstract
Background
RNase III is a dsRNA specific endoribonuclease which is involved in the primary processing of rRNA and several mRNA species in bacteria. Both primary structural elements and the secondary structure of the substrate RNA play a role in cleavage specificity.
Results
We have analyzed RNase III cleavage sites around both ends of pre-23 S rRNA in the ribosome and in the protein-free pre-rRNA. It was found that in the protein-free pre-23 S rRNA the main cleavage site is at position (-7) in respect of the mature 5' end. When pre-23 S rRNA was in 70 S ribosomes or in 50 S subunits, the RNase III cleavage occurred at position (-3). We have demonstrated that RNase III interacts with both ribosomal subunits and with even higher affinity with 70 S ribosomes. Association of RNase III with 70 S ribosomes cannot be dissociated by poly(U) RNA indicating that the binding is specific.
Conclusions
In addition to the primary and secondary structural elements in RNA, protein binding to substrate RNA can be a determinant of the RNase III cleavage site.
Background
In Escherichia coli, each of the seven rRNA operons is transcribed as a 30 S precursor containing functional gene products – 16 S, 23 S, 5 S and tRNA(s). In addition, 30 S precursor molecules contain extra sequences in the beginning and end of structural genes and in between, as well. These additional sequences are removed by a series of processing events [1]. Enzymes involved in rRNA processing have been the subject of extensive studies, but a full understanding of this important process is still incomplete [reviewed in [1,2]]).
RNase III is the best studied enzyme involved in rRNA processing. RNase III is highly conserved in the eubacteria and orthologs occur in fungi, plants and animals [3,4]. In E. coli RNase III is encoded by the rnc gene [5], which maps at 55 minutes. It is a double-stranded RNA specific enzyme [6], which is active as a 52 kDa homodimer [7] and requires a divalent metal ion (preferably Mg2+) for activity [8]. In addition to the secondary structure of substrate RNA, both positive (determinants) and negative (antideterminants) sequence elements are important in determining the cleavage site [2].
In Escherichia coli, sequences flanking both 16 S RNA and 23 S RNA contain complementary sequence tracts that can form double-stranded regions enclosing the sequences of the mature species of 16 S and 23 S [9]. These helices are known as processing stems. As a result of RNase III cleavage, pre-16 S and pre-23 S precursor molecules are released [10]. This event has been shown to occur during transcription of rRNA [11]. Only 1–2% of rRNA is present as long precursors in wild-type bacteria [12]. The final maturation of rRNA molecules depends on the formation of the complex of rRNA and r-proteins and requires participation of other ribonucleases [1].
Bacteria lacking RNase III activity are viable, although slow growth and defective translation of some specific mRNA's has been attributed to the lack of the enzyme [11]. In these strains, other ribonucleases are responsible for the formation of 16 S and 23 S precursors. However, these precursors are somewhat larger than the precursors formed by RNase III cleavage [13]. Unlike 16 S rRNA, the mature ends of 23 S rRNA are not formed in the RNase III-negative strains [1].
RNase III processing sites in the 23 S precursor have been localized at 3 and 7 nucleotides upstream from the mature 23 S rRNA 5' terminus [14,15] and at 8 nucleotides downstream from the 3' terminus [16]. In 23 S rRNA, initial cleavage by RNase III is indispensable for maturation, and no mature 23 S RNA is found in RNase III¯ strains [16]. The mature 5' terminus of 23 S rRNA is formed in ribosomes by an unknown enzyme. This seems to be endoribonuclease, because no intermediates have been observed. The maturation of the 3' terminus requires RNase T [17]. RNase III was tought to cleave both strands of the 23 S processing stem simultaneously in a staggered fashion. However, more recently it has been demonstrated, that the maturation of the 5' end of 23 S rRNA occurs independently of that at the 3' terminus [17]. Interestingly, there is a reciprocal dependency between RNase III activity and ribosome assembly, the mature 5' end of 16 S rRNA and both mature ends of 23 S rRNA are known to be formed only in assembled ribosomes [1]. On the other hand, a temperature-sensitive mutation in ribosomal protein S12, which affects small subunit assembly, has been found to be suppressed by a mutation in RNase III [18].
In the present work we have compared RNase III cleavage sites on different forms of precursor 23 S rRNA in vitro. It was found that RNase III cleaves pre-23 S rRNA in ribosomes at position 3 nucleotides upstream from the mature 5' terminus of 23 S rRNA. Deproteinized rRNA from the rnc- strain and in vitro transcribed rRNA were cleaved by RNase III at position (-7). Thus, the specificity of RNase III cleavage is altered by the presence of ribosomal proteins. We have also shown that RNase III binds to the ribosomes. Binding to the 70 S ribosomes is stronger when compared to the dissociated subunits.
Results and discussion
In order to analyze the possible link between ribosome assembly and RNase III processing events we have isolated ribosomes from RNase III negative and from the wild-type strains. In the absence of RNase III the extended version of 23 S rRNA accumulates in the ribosome [9,16]. The primer extension identified most of the 5' ends of 23 S rRNA at positions (-42) – (-45) (data not shown). However, a significant fraction of the 5' ends were located in the helix 1 region of mature 23 S rRNA (positions 3–6). These results are in agreement with the earlier observations [15,16].
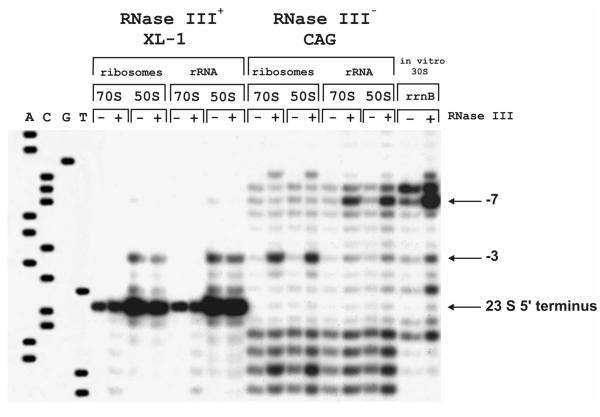
Pre-23 S rRNA was cleaved by RNase III in the ribosomes and after removal of ribosomal proteins. After treatment of 70 S ribosomes or 50 S subunits with RNase III, the main 5' end of 23 S rRNA was at position (-3) (Fig. 1). In contrast, if ribosomal proteins were removed by phenol extraction, the main RNase III cleavage on the naked pre-23 S rRNA occurred at position (-7), although very little cleavage also occurred at position (-3). The in vitro transcribed, unmodified 30 S rRNA precursor from the rrnB operon was cleaved by RNase III preferentially at position (-7) (Fig. 1). These results clearly demonstrate that the specificity of RNase III is altered by the presence of ribosomal proteins. It is interesting to note that the RNase III cleavage site in the ribosomes (-3) is closer to the ribosomal body when compared to the cleavage site in the free rRNA (-7). Therefore, the presence of r-proteins in the 50 S subunit does not protect or mask the distal cleavage site. We assume that binding RNase III to the ribosomes directs the cleavage at position (-3).
Figure 1.
RNase III cleavage sites in the 5' strand of pre-23 S rRNA. 70 S ribosomes and 50 S subunits were isolated from the RNase III positive (XL-1B) and from RNase III negative (CAG) strain. The 30 S rRNA precursor was transcribed in vitro. Samples were treated with RNase III (+ lanes). Control samples were incubated without the enzyme. RNase III cleavage sites on the 23 S rRNA in the ribosomes or in the naked RNA were determined by a reverse transcriptase primer extension. The sequence of the rrnB operon around the 5' end of 23 S rRNA is shown on lines A, C, G, T. RNase III cleavage sites and the mature 5' end of 23 S rRNA are indicated on the right (-7, -3, and 5' 23S).
Next, we have analyzed the 5' end of 23 S rRNA in the 70 S ribosomes and in the 50 S subunits of the wild-type strain (RNase III positive strain; see Fig. 1). Only the mature 5' end was found in the 70 S ribosomes, while the 50 S subunits contain a significant fraction of 5' ends at position (-3). Final maturation of 23 S rRNA is known to occur in 70 S ribosomes or in polysomes [14]. Thus, the 50 S subunits with extended 23 S rRNA represent newly formed 50 S subunits which have not been in 70 S. Moreover, the fact that in the 50 S subunits 23 S rRNA has a 5' end at position (-3) suggests that RNase III cleavage occurs after the 50 S subunits are assembled. Srivastava and Schlessinger have proposed that RNase III cleavage of pre-rRNA occurs on the level of ribonucleoprotein particles [1]. Our results confirm this. The fact that rRNA cleavage by RNase III happens after the r-proteins are associated with the 23 S rRNA supports the view that ribosomal large subunit assembly is coupled to the transcription of 23 S rRNA [19].
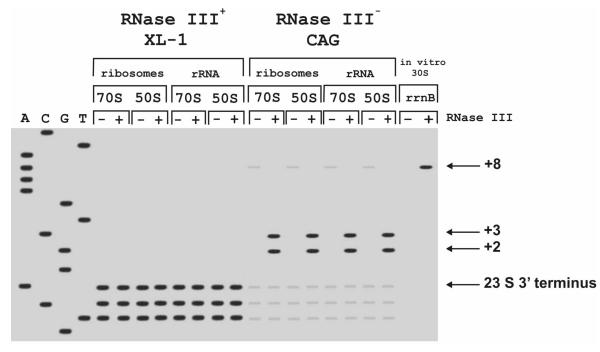
In similar experiments we have analyzed RNase III cleavage sites on the 3' strand of pre-23 S rRNA. The cleavage sites were determined by S1 nuclease mapping. In the wild-type strain, the 3' end of 23 S rRNA was heterogeneous, in addition to the known 3' end the molecules one or two nucleotides shorter were also found (Fig. 2). Similar heterogeneity at the 3' end of 23 S rRNA was also observed by others (e. g. in [17]). When ribosomes were isolated from the wild-type, RNase III treatment did not have any effect on the 3' end of 23 S rRNA either in the ribosomes or in the free rRNA.
Figure 2.
RNase III cleavage sites in the 3' strand of pre-23 S rRNA. 70 S ribosomes, 50 S subunits or in vitro transcribed 30 S pre-rRNA were incubated with or without RNase III (+ and - lanes respectively). The cleavage sites were determined by S1 nuclease mapping. Sequencing lanes A, C, G, and T were used to determine of exact length of protected 3' [32P]DNA. The positions of the RNase III cleavage and the mature 3' end of 23 S rRNA are indicated on the right.
50 S subunits isolated from the RNase III-negative strain CAG1632 contain extensions 43, 44, and <45 nucleotides at the 3' end (data not shown). 70 S ribosomes and 50 S subunits, isolated from RNase III-negative strain, were treated by RNase III. Two cleavage sites corresponding to positions (+2) and (+3) in respect of the canonical 3' end of 23 S rRNA were identified by S1 mapping (Fig. 2). The same cleavage sites were found when the ribosomal proteins were removed prior to the RNase III cleavage reaction. Thus, in contrast to the RNase III cleavage at the 5' end of pre-23 S rRNA, the cleavage at the 3' end is not influenced by the presence of the ribosomal proteins.
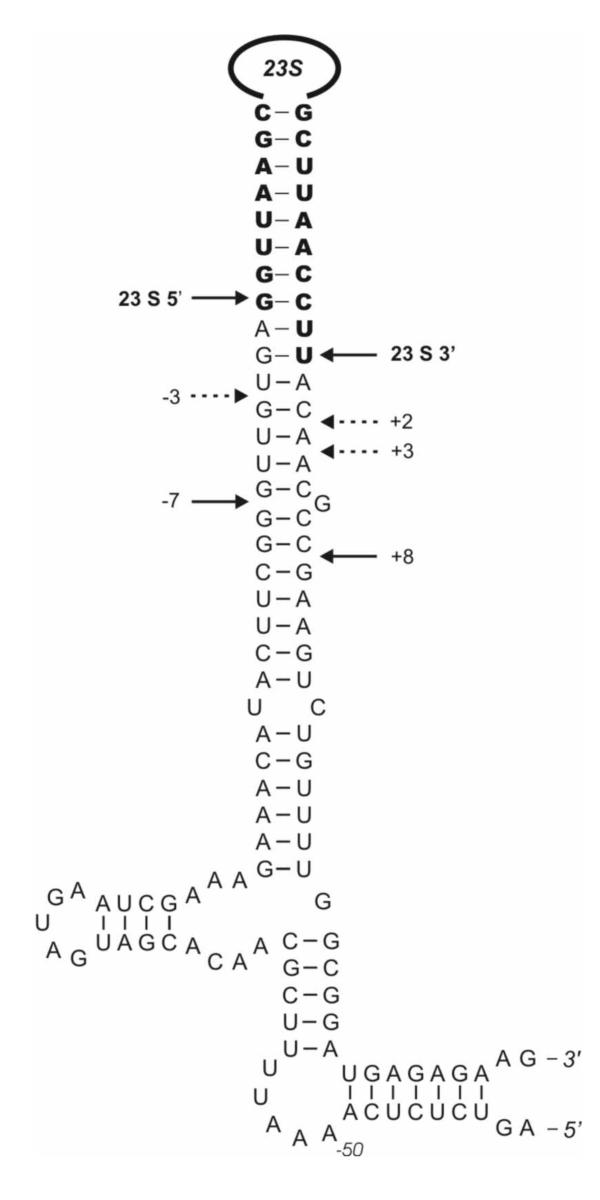
On the in vitro transcribed rrnB 30 S precursor RNase III cleaves the 23 S rRNA 3' strand at position (+8). This site has been recognized before as the RNase III cleavage site [16]. The results presented here suggest that the RNase III cleavage site before ribosomal assembly is at position (+8). When 50 S subunits are formed, the RNase III cleaves at positions (+2) and (+3). RNase III cleavage sites on the processing stem of pre-23 S rRNA are summarized in Fig. 3.
Figure 3.
Secondary structure of the end region of pre-23 S rRNA (processing stem). Mature 23 S rRNA is shown in bold. The 5' and 3' ends and the RNase III cleavage sites on the naked pre-23 S rRNA are indicated by solid lines and the cleavage sites in the ribosomes by dotted lines.
Since ribosomes alter the specificity of RNase III cleavage, we tested whether RNase III can bind to 70 S ribosomes or to the ribosomal subunits. RNase III has been shown to copurify with ribosomes [20] suggesting physical interaction with the ribosomes. Radioactive RNase III was incubated with ribosomes and passed through the Sephacryl S400 spin column. Approximately 60–80% of the 70 S ribosomes are found in the void volume. 12–15% of the radioactivity was also present in the ribosomal fraction, while less than 1% of the radioactivity was found in this fraction in the absence of added ribosomes (Table 1). This result indicates significant binding of RNase III to the 70 S ribosomes. The actual binding of RNase III to the 70 S ribosomes can be higher since 20–40% of the ribosomes remain in the column. Similar results in RNase III binding were detected with ribosomes from the RNase III-negative strain and the wild type strain.
Table 1.
Association of RNase III with ribosomes and ribosomal subunits.
| dpm | |
| 70 S (CAG1632) | 4507 ± 379 |
| 70 S (XL-1B) | 3688 ± 345 |
| 50 S | 1241 ± 97 |
| 30 S | 1274 ± 141 |
| poly(U) | 541 ± 36 |
| 70 S + poly(U) | 2511 ± 239 |
| - | 179 ± 34 |
[35S]RNase III (30 000 dpm) was incubated with 1 A260 unit 70 S ribosomes or with the same amount of isolated subunits followed by separation of ribosomes from unbound radioactivity by a Sephacryl S400 spin column using NH4Cl concentration as indicated. Ribosome-bound RNase III was determined by TCA precipitation and counting the precipitate for radioactivity. The results are shown in dpm. In the control experiments 2 A260 units of poly(U) was added to the incubation mixture. The control with [35S]RNase III alone is also shown. The data presented here are average results of 4 parallel experiments.
Significant binding of RNase III to both 50 S and 30 S subunits was detected (Table 1). Taken together, RNase III binds to the 70 S ribosomes and to both subunits. However, RNase III has a higher affinity to the 70 S ribosomes when compared to the isolated subunits. RNase III is known to bind dsRNA via its C-terminal dsRNA binding domain [21]. It is therefore reasonable to assume that RNase III interacts with the rRNA. The fact that ribosomal proteins alter the specificity of RNase III cleavage reaction suggests direct interaction between RNase III and r-proteins. Thus, both rRNA and r-proteins are likely to participate in RNase III – ribosome interaction.
To further test the specificity of RNase III binding to the ribosomes, we have performed competition experiments. Polyuridylic acid (average 4 S) was used as unspecific competitor RNA. When RNase III was incubated with 2 A260 units of poly(U) and passed through the Sephacryl S400 spin column a small amount of RNase III was found in the void volume (Table 1). Poly(U) is apparently not big enough to be in the flow-through fraction. Thus, in the presence of poly(U) RNase III remains in the column. When RNase III binding to 70 S ribosomes (1 A260 unit) was tested in the presence of 2 A260 units poly(U) 5 times more RNase III was found in the ribosome fraction when compared to the poly(U) binding results. Therefore, poly(U) did not efficiently compete with the ribosome for the RNase III binding. We conclude that the binding to the ribosomes is specific.
The results described above demonstrate that RNase III binds to both isolated ribosomal subunits and with an even higher affinity to the 70 S ribosomes. Such interaction can be important for ribosome biogenesis to facilitate high molecular rRNA processing. The same idea has been published by A. Nicholson [2]. On the other hand, the translation of many mRNAs in E. coli is regulated by RNase III cleavage [2]. If translation of an mRNA is coupled to transcription and is regulated by RNase III, the association of the enzyme with the ribosome would enhance the control of translation by RNase III. An example of RNase III regulated mRNA is transcription antitermination factor N of phage λ. Recently it has been demonstrated that RNase III cleavage does not inhibit gene N transcription antitermination but prevents N-mediated translation repression of N gene expression [22]. In this system, the association of RNase III with the 70 S ribosomes can play a role in regulating gene expression at the level of translation. It remains to be tested whether the RNase III associated with 70 S has the same cleavage specificity as the free enzyme.
Conclusions
1. The specificity of RNase III is altered by the presence of ribosomal proteins.
2. RNase III binds to both the isolated ribosomal subunits and with an even higher affinity to the 70 S ribosomes.
Materials and methods
Strains and plasmids
E. coli strains XL1-Blue (supE44 hsdR17 recA1 endA1 gyrA46 thi relA1 lac- F'(proAB+lacIqLacZΔM15 Tn10(tetr)) and rnc--deficient strain CAG1632 (F- his bio RNase I- RNase III- relA recA56 srl::Tn10; [23]) were used for isolation of the ribosomes.
Modified strain BL'21 (DE3) carrying plasmid pET-15b [rnc-wt] was kind gift from Dr. A. Nicholson (Detroit, USA). Originally RecA strain was changed into RecA- and contains a point mutation in the chromosomal rnc gene, causing the synthesis of catalytically inactive RNase III from the chromosome.
Plasmid pET-15b [rnc-wt] contains rnc gene encoding RNase III between BamHI and NdeI sites under the control of the late promoter of T7 bacteriophage. Plasmid pAR-Xho containing rrnB operon with XhoI site downstream of 5S gene was a kind gift from Dr. Rolf Wagner (University of Düsseldorf, Germany).
Purification of RNase III
RNase III-His6 was expressed in BL'21 (DE3)/pET-15b [rnc-wt] in 2xYT medium. The enzyme was isolated using 1 ml of Ni-NTA gel (Quiagen). Protein was eluted with 6 × 0,5 ml elution buffer (50 mM NaH2PO4; 300 mM NaCl; 250 mM imidazol). Collected fractions were tested for their enzymatic activity and fractions containing activity were dialyzed against the storage buffer (50% glycerol; 0,5 M KCl; 30 mM Tris pH 6.3; 0,1 mM EDTA; 0,1 mM DTT).
[35S]RNase III-His6 was expressed in the M9 minimal medium. Cells were induced at A600 = 0,2 by IPTG (1 mM final concentration). After 20 minutes 0,5 mCi [35S] was added. Cells were collected after 8 hours. RNase III was isolated as described above. Functional activity of the RNase III was tested according to the ability to process in vitro transcribed 30 S precursor into pre-16 S and pre-23 S rRNA.
Transcription in vitro
Plasmid pAR-Xho was used for transcription of 30 S precursor corresponding to rrnB. pAR-Xho was linearized by Xho I. Linearized plasmid was purified form the mixture using UltraClean™ 15 kit (MoBio Laboratories).
Transcription was carried out in 50 μl buffer (50 mM NH4Cl; 40 mM Tris, pH 7,9; 20 mM MgCl2; 2 mM spermidine; 1 mM DTT), in the presence of 2,5 mM rNTP, 0,1 μg linear template DNA, 20 units RNasine and T7 RNA polymerase. Reaction was incubated at 37°C for 1 hour in the presence of optimized amount of RNase III or wihout the enzyme.
Ribosomes and rRNA
XL-1 Blue and CAG1632 cells were grown in 2xYT medium up to A600 = 0.9. Cells were lysed and ribosomes fractionated by sucrose gradient (10–25%) centrifugation in SW-28 by ω2t = 2,8 × 1011 as described previously [15]. Ribosomes were collected by ethanol precipitation. Ribosomes were deproteinized by phenol and chloroform extractions followed by ethanol precipitation of rRNA.
Mapping of 5' ends
RNase III cleavage sites around the 5' end of 23 S rRNA were identified by reverse transcriptase primer extension using the primer 5'-CGCCTCTGACTGCCAGGGC-3' (complementary to the bases 3529–3547 of rrnB operon, numbering according to [24]) as described [15]. AMV reverse transcriptase was from Seikagaku (Japan). The same primer was used for DNA sequencing by T7 sequencing kit (Amersham Pharmacia Biotech).
Mapping of 3' ends
RNase III cleavage sites at the 3' side of pre-23 S rRNA were determined by S1 mapping. Single stranded DNA complementary to the positions 6374–6540 (numbering according to [24]) was labelled by [α32P]-ddATP using terminal transferse (Fermentas, Lithuania) according to manufacturers recommendations. ccDNA probe (1 pmol) was mixed with 4 pmoles of rRNA and hybridized in 20 μl buffer H (0.1 M HEPES, pH 7,8; 1 mM EDTA; 0.5 M NaCl) at 65°C for 20 min. 300 μl of buffer S (280 mM NaCl; 4.5 mM ZnSO4; 50 mM NaOAc pH 4.5; herring sperm DNA 20 μg/ml) and 10 units S1 nuclease (Fermentas, Lithuania) were added. Incubation was 40 min. at 37°C. Reaction was stopped by EDTA/NaCl and phenol extraction of DNA. Products were separated by 10% urea-PAGE.
RNase III binding to the ribosomes
Binding of RNase III to the 70 S ribosomes and ribosomal subunits was tested by spin column method. [35S]RNase III (30 000 dpm) was incubated with 1 A260 unit ribosomes 10 min. on ice in 50 μl buffer B (10 mM MgCl2; 20 mM Tris-HCl ph 7.6; 6 mM 2-ME; 100 mM NH4Cl). Ribosome-bound RNase III was separated from free enzyme by 1 ml Sephacryl S-400 (Amersham Pharmacia Biotech) spin column equilibrated by the buffer B with corresponding NH4Cl concentration. 5% TCA precipitate was collected and counted for radioactivity. In the absence of ribosomes 100–200 dpm was found in the flowthrough fraction.
Acknowledgments
Acknowledgments
We thank our colleagues for help and advise, MSc Ü. Maiväli (Tartu University) for critically reading the manuscript. This work was made possible by Howard Hughes Medical Institute International Research Grant 55000332 and was supported by Estonian Science Foundation Grant 4425.
Contributor Information
Ülar Allas, Email: snoopy@ut.ee.
Aivar Liiv, Email: aliiv@ebc.ee.
Jaanus Remme, Email: jremme@ebc.ee.
References
- Srivastava AK, Schlessinger D. Mechanism and regulation of bacterial ribosomal RNA processing. Annu Rev Microbiol. 1990;44:105–129. doi: 10.1146/annurev.micro.44.1.105. [DOI] [PubMed] [Google Scholar]
- Nicholson AW. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol Rev. 1999;23:371–390. doi: 10.1016/S0168-6445(99)00013-3. [DOI] [PubMed] [Google Scholar]
- Rotondo G, Frendewey D. Purification and characterization of the Pac1 ribonuclease of Schizosaccharomyces pombe. Nucleic Acids Res. 1996;24:2377–2386. doi: 10.1093/nar/24.12.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Xu H, Miraglia LJ, Crooke ST. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J Biol Chem. 2000;275:36957–36965. doi: 10.1074/jbc.M005494200. [DOI] [PubMed] [Google Scholar]
- Nashimoto H, Uchida H. DNA sequencing of the Escherichia coli ribonuclease III gene and its mutations. Mol Gen Genet. 1985;201:25–29. doi: 10.1007/BF00397981. [DOI] [PubMed] [Google Scholar]
- Robertson HD, Webster RE, Zinder ND. Purification and properties of ribonuclease III from Escherichia coli. J Biol Chem. 1968;243:82–91. [PubMed] [Google Scholar]
- Dunn JJ. RNase III cleavage of single-stranded RNA. Effect of ionic strength on the fideltiy of cleavage. J Biol Chem. 1976;251:3807–3814. [PubMed] [Google Scholar]
- Dunn JJ. Ribonuclease III. In: Boyer PD, editor. The Enzymes. Vol. 15. New York, Academic Press; 1982. pp. 485–499. [Google Scholar]
- Bram RJ, Young RA, Steitz JA. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980;19:393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- Nikolaev N, Schlessinger D, Wellauer PK. 30 S pre-ribosomal RNA of Escherichia coli and products of cleavage by ribonuclease III: length and molecular weight. J Mol Biol. 1974;86:741–747. doi: 10.1016/0022-2836(74)90350-7. [DOI] [PubMed] [Google Scholar]
- Apirion D, Gegenheimer P. Processing of RNA. Boca Raton Fla: CRC Press; 1984. Molecular biology of RNA processing in procaryotic cells; pp. 36–52. [Google Scholar]
- King TC, Schlessinger D. S1 nuclease mapping analysis of ribosomal RNA processing in wild type and processing deficient Escherichia coli. J Biol Chem. 1983;258:12034–12042. [PubMed] [Google Scholar]
- Gegenheimer P, Watson N, Apirion D. Multiple pathways for primary processing of ribosomal RNA in Escherichia coli. J Biol Chem. 1977;252:3064–3073. [PubMed] [Google Scholar]
- Sirdeshmukh R, Schlessinger D. Ordered processing of Escherichia coli 23S rRNA in vitro. Nucleic Acids Res. 1985;13:5041–5054. doi: 10.1093/nar/13.14.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liiv A, Remme J. Base-pairing of 23 S rRNA ends is essential for ribosomal large subunit assembly. J Mol Biol. 1998;276:537–45. doi: 10.1006/jmbi.1997.1532. [DOI] [PubMed] [Google Scholar]
- King TC, Sirdeshmukh R, Schlessinger D. RNase III cleavage is obligate for maturation but not for function of Escherichia coli pre-23S rRNA. Proc Natl Acad Sci USA. 1984;81:185–188. doi: 10.1073/pnas.81.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Pandit S, Deutscher MP. Maturation of 23S ribosomal RNA requires the exoribonuclease RNase T. RNA. 1999;5:139–146. doi: 10.1017/S1355838299981669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashimoto H, Miura A, Saito H, Uchida H. Suppression of temperature-sensitive mutations in ribosomal protein gene, rpsl (S12), of Escherichia coli K12. Mol Gen Genet. 1985;199:381–387. doi: 10.1007/BF00330746. [DOI] [PubMed] [Google Scholar]
- Lewicki BT, Margus T, Remme J, Nierhaus KH. Coupling of rRNA transcription and ribosomal assembly in vivo. Formation of active ribosomal subunits in Escherichia coli requires transcription of rRNA genes by host RNA polymerase which cannot be replaced by bacteriophage T7 RNA polymerase. J Mol Biol. 1993;231:581–593. doi: 10.1006/jmbi.1993.1311. [DOI] [PubMed] [Google Scholar]
- Robertson HD, Webster RE, Zinder ND. Purification and properties of ribonuclease III from Escherichia coli. J Biol Chem. 1968;243:82–91. [PubMed] [Google Scholar]
- Kharrat A, Macias MJ, Gibson TJ, Nilges M, Pastore A. Structure of the dsRNA binding domain of E. coli RNase III. EMBO J. 1995;14:3572–3584. doi: 10.1002/j.1460-2075.1995.tb07363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson HR, Yu D, Peters HK, 3rd, Zhou JG, Court DL. The global regulator RNase III modulates translation repression by the transcription elongation factor N. EMBO J. 2002;21:4154–4161. doi: 10.1093/emboj/cdf395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory ST, O'Connor M, Dahlberg AE. Functional Escherichia coli 23S rRNAs containing processed and unprocessed intervening sequences from Salmonella typhimurium. Nucleic Acids Res. 1996;24:4918–4923. doi: 10.1093/nar/24.24.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J, Dull TJ, Noller HF. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1980;77:201–204. doi: 10.1073/pnas.77.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]