Abstract
The MICdb (Microsatellites Database) (http://www.cdfd.org.in/micas) is a comprehensive relational database of non-redundant microsatellites extracted from fully sequenced prokaryotic genomes. The current version (1.0) of the database has been compiled from 83 genomes belonging to different phylogenetic groups. This database has been linked to MICAS, the web-based Microstatellite Analysis Server. MICAS provides a user-friendly front-end to systematically extract data on microsatellite tracts from genomes. The database contains the following information pertaining to the microsatellites: the regions (coding/non-coding, if coding, their GenBank annotations) containing microsatellite tracts; the frequencies of their occurrences, the size and the number of repeating motifs; and the sequences of the tracts. MICAS also provides an interface to Autoprimer, a primer design program to automatically design primers for selected microsatellite loci.
INTRODUCTION
Microsatellites, also known as simple sequence repeats, are short, tandem repeats of 1–6 nt occurring in most of the genomes. They serve as excellent molecular markers for genotyping, strain differentiation, epidemiological analysis and genome analysis (1–3). These elements also play very important roles in phase variation of pathogenic bacteria by regulating genes and gene products (4–10). Microsatellite markers have also been proven to be rapid tools for identifying pathogenic bacteria from clinical isolates (11,12).
Availability of complete and annotated genome sequences of a number of organisms has provided an excellent opportunity to analyse microsatellites in a very great detail for their genomic locations, distributions and frequencies. Results from such analysis provide a useful basis for carrying out further investigations into the structural and functional characteristics of microsatellites. During the course of such investigations we developed a fully automated software for locating microsatellites in a given sequence (VB Sreenu, J Nagaraju and HA Nagarajaram, manuscript under preparation). Using this software we carried out systematic searches and extracted non-redundant microsatellites from the sequences of 83 different organisms and stored them in the form of a relational database called MICdb (Microsatellites Database). In this communication we provide a brief description of this database and its utility.
STRUCTURE OF THE DATABASE
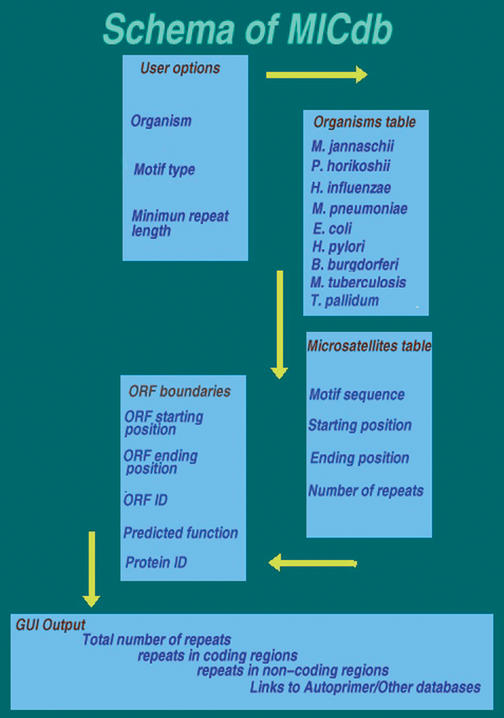
MICdb has been developed using MySQL (www.mysql.com). The information stored in the database includes genomic location of microsatellites (starting and ending positions), the motif types (mono, di, etc.), the sequences of the motifs, regions of occurrence (coding, non-coding, etc.) and frequencies of occurrence in the entire genome. The information pertaining to the coding regions such as the gene identifier, description of protein function etc. are also included. Currently the database comprises of 913 tables (83×11 tables) i.e. 11 tables per genome. Of the 11 tables for a genome, the first 10 contain information pertaining to repeats of size, mono to deca, respectively (in addition to motif length mono to hexa, the longer motifs of length 7 to 10 are also included). The eleventh table contains information on the coding regions (see Fig. 1). Tables holding information about microsatellites from mono to deca are identical in their structure comprising of six fields (Table 1A). The seventh table where ORF information is stored also has six fields (Table 1B). Schema of MICdb and flow of the data are illustrated in Figure 1.
Figure 1.
Schema of MICdb and data flow.
Table 1A. Model of MySQL table which is used for storing microsatellites information.
| Field | Type | Null | Key | Default | Extra |
|---|---|---|---|---|---|
| Motif | varchar (15) | YES | NULL | ||
| Repeat | int (2) | YES | NULL | ||
| Sp | int (11) | YES | NULL | ||
| Ep | int (11) | YES | NULL | ||
| Region | char (1) | YES | NULL | ||
| Strand | char (1) | YES | NULL |
First field (Motif) is for storing motif sequence.
Second field (Repeat) is for repeat length.
Third field (Sp) is for starting position of repeat.
Fourth field (Ep) is for ending position of repeat.
Fifth field (Region) for coding and non-coding information.
Sixth field (Strand) for coding strand (+ or −).
Table 1B. Model of MySQL table which is used for storing information pertaining to coding regions.
| Field | Type | Null | Key | Default | Extra |
|---|---|---|---|---|---|
| PROT_ID | varchar (50) | YES | NULL | ||
| PROT_DESC | varchar (255) | YES | NULL | ||
| ORF_ID | varchar (200) | YES | NULL | ||
| STRAND | char (1) | YES | NULL | ||
| ORF_SPOS | int (11) | YES | NULL | ||
| ORF_EPOS | int (11) | YES | NULL |
First field (PROT_ID) is for gene identifier.
Second field (PROT_DESC) is for protein description (function).
Third field (ORF_ID) is for ORF identification number.
Fourth field (STRAND) is coding strand information (+ or −).
Fifth field (ORF_SPOS) is for ORF starting position.
Sixth field (ORF_EPOS) is for ORF ending position.
DATA EXTRACTION
A web-interface to MICdb has been provided with the help of a server called MICAS (Microsatellite Analysis Server) which provides an user-friendly front-end to the database for data retrieval. In order to query the database for a microsatellite the user has to first select a genome followed by the motif size (S) and the repeat number (N). MICAS retrieves all the microsatellite tracts made up of the motifs of size S repeating at least N number of times in the genome. The retrieved results are displayed in the form of a table which contains the sequences of the repeating units, the minimum and maximum number of times the units are found repeated at different loci and the frequency of their occurrence in the entire genome. The user can select a tract and query the database for further details. These details are the starting and ending positions of the microsatellite tracts, the region in which the tract occurs, coding or non-coding and if coding, function of the translated product and strand (+/−) in which the coding occurs. The coding regions are hyperlinked texts linked to the annotated information deposited in GenBank. Further, the table also provides a link to the Autoprimer software for every microsatellite tract. Autoprimer is a primer design software developed by us to design primers for a selected nucleotide tract containing microsatellite. Autoprimer takes care of repeat regions in the primers, checks for self-complimentarity and primer pair complimentarity by using dynamic programing. The software uses the nearest neighbour method (13) for calculating melting temperatures (Tm). A user can click the link by which MICAS initiates automatically the Autoprimer input page which contains the full sequence of the microsatellite along with flanking regions of default size (100 bp) and the criteria (melting temperature, GC content etc.) for primer design and selection. Users can change these criteria. The output from Autoprimer is a list of optimally designed primers.
FUTURE PERSPECTIVES
MICdb is committed to provide the scientific community with comprehensive information on microsatellites occurring in all the published, publicly available genomes. MICdb is upgraded regularly. Currently MICdb contains information extracted from 83 prokaryotic genomes. As the database creation has been made fully automated the database can be updated for any number of genomes. Presently the database has a hyperlink only to GenBank for downloading the annotated information pertaining to the coding regions of the genomes. In the future version, hyperlinks to other useful databases will also be provided thereby increasing the information content associated with the microsatellites.
AVAILABILITY
MICdb is accessible via the World Wide Web interface at http://www.cdfd.org.in/micas. The site has been designed to include a user friendly navigation system and more graphical interfaces and analysis tools like MICAS and Autoprimer. The present article reflects the up-to-date upgradation of the database and should be cited accordingly.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Miss Sushma for assisting in the design of the Autoprimer software. V.B.S. gratefully acknowledges the Council of Scientific and Industrial Research (CSIR), Government of India, for the Junior Research Fellowship. H.A.N. and J.N. gratefully acknowledge the core-grant from CDFD and an extramural grant from the Department of Biotechnology (DBT), Government of India, respectively.
REFERENCES
- 1.Van Soolingen D., de Haas,P.E.W., Heramans,P.W.M., Groenen,P.M.A. and van Embden,J.D.A. (1993) Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J. Clin. Microbiol., 31, 1987–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gur-Arie R., de Haas,P.E.W., Heramans,P.W.M., Groenen,P.M.A. and van Embden,J.D.A. (1993) Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J. Clin. Microbiol., 31, 1987–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen G.L., de Haas,P.E.W., Heramans,P.W.M., Groenen,P.M.A. and van Embden,J.D.A. (1993) Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J. Clin. Microbiol., 31, 1987–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borst P., de Haas,P.E.W., Heramans,P.W.M., Groenen,P.M.A. and van Embden,J.D.A. (1993) Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J. Clin. Microbiol., 31, 1987–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burch C.L., de Haas,P.E.W., Heramans,P.W.M., Groenen,P.M.A. and van Embden,J.D.A. (1993) Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J. Clin. Microbiol., 31, 1987–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hood D.W., de Haas,P.E.W., Heramans,P.W.M., Groenen,P.M.A. and van Embden,J.D.A. (1993) Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J. Clin. Microbiol., 31, 1987–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makino S., de Haas,P.E.W., Heramans,P.W.M., Groenen,P.M.A. and van Embden,J.D.A. (1993) Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J. Clin. Microbiol., 31, 1987–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy G.L., de Haas,P.E.W., Heramans,P.W.M., Groenen,P.M.A. and van Embden,J.D.A. (1993) Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J. Clin. Microbiol., 31, 1987–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peak I.R.A., de Haas,P.E.W., Heramans,P.W.M., Groenen,P.M.A. and van Embden,J.D.A. (1993) Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J. Clin. Microbiol., 31, 1987–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roche R.J., de Haas,P.E.W., Heramans,P.W.M., Groenen,P.M.A. and van Embden,J.D.A. (1993) Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J. Clin. Microbiol., 31, 1987–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marshall D.G., de Haas,P.E.W., Heramans,P.W.M., Groenen,P.M.A. and van Embden,J.D.A. (1993) Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J. Clin. Microbiol., 31, 1987–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Belkum A., de Haas,P.E.W., Heramans,P.W.M., Groenen,P.M.A. and van Embden,J.D.A. (1993) Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J. Clin. Microbiol., 31, 1987–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breslauer K.J., de Haas,P.E.W., Heramans,P.W.M., Groenen,P.M.A. and van Embden,J.D.A. (1993) Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J. Clin. Microbiol., 31, 1987–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]