Abstract
Rfam is a collection of multiple sequence alignments and covariance models representing non-coding RNA families. Rfam is available on the web in the UK at http://www.sanger.ac.uk/Software/Rfam/ and in the US at http://rfam.wustl.edu/. These websites allow the user to search a query sequence against a library of covariance models, and view multiple sequence alignments and family annotation. The database can also be downloaded in flatfile form and searched locally using the INFERNAL package (http://infernal.wustl.edu/). The first release of Rfam (1.0) contains 25 families, which annotate over 50 000 non-coding RNA genes in the taxonomic divisions of the EMBL nucleotide database.
INTRODUCTION
Non-coding RNA genes produce a functional RNA molecule as a final product, rather than a translated protein. Current gene-finding methods largely ignore non-coding RNA genes, yet they produce some of the cell's most important products—transfer RNA and ribosomal RNA are two of the well-known examples. The number of known RNA genes is expanding rapidly due to the deluge of genomic data, but also aided by recent systematic efforts to detect RNA genes [reviewed in (1–3)].
Just like protein coding genes ncRNAs fall into families that have evolved from a common ancestor. By making alignments of these families of ncRNA genes we can learn about their structure and function. Indeed, accurate prediction of RNA secondary structure relies on multiple sequence alignments to provide data on co-varying bases (4). Ribosomal RNA alignments are used to make molecular phylogenies that guide taxonomic classification of all species (5).
Many RNA sequence families conserve a consensus base-paired secondary structure. Standard primary sequence analysis tools [such as BLAST (6) for database searches and CLUSTALW (7) for multiple alignment] are useful for closely related RNAs, but recognition and alignment of more distantly related structural RNAs is greatly aided by consensus secondary structure information. Historically, structure-based RNA sequence analysis has been difficult to automate. Most RNA structural alignments are the product of expert manual curation. Recent software advances (8) using secondary structure profiles called ‘covariance models’ (CMs—also called profile stochastic context-free grammars) (1,9) have led us to begin the development and automated maintenance of a database of structural RNA alignments. This is analogous to the use of profile hidden Markov models of primary sequence consensus in the development and maintenance of thousands of protein sequence alignments in the Pfam database (10).
Several databases already exist that contain RNA alignments and information—for example, the European Large Subunit Ribosomal RNA Database (11), the SRP database (12), the uRNA database (13), the Comparative RNA Web (14), and others (15–22). These databases are well curated and provide a large amount of information to the specialist. However, they vary greatly in the file formats used and the data presented. There are also several specialised computational tools to aid identification of specific RNA types. For example, tRNAscan-SE is a standard tool in the genome annotation field for identifying tRNA genes with extremely high sensitivity and specificity (23). A recent report describes a new tool, BRUCE, which aims to predict tmRNA genes in genomic sequence (24). However, the RNA analysis field lacks any analogue to the comprehensive secondary sequence databases that greatly aid protein annotation, such as Pfam (10), SMART (25) and Prosite (26).
The aims of the Rfam database are (i) to integrate the many existing curated structural RNA alignments (in addition to new alignments) into a common structure-annotated format, analogous to Pfam's curated seed alignments; (ii) to use covariance model software to search the growing sequence databases and maintain automatically-generated alignments of all detectable homologues, analogous to Pfam's automatically-generated full alignments; and (iii) to provide a system for automatically analysing and annotating sequences (including complete genome sequences) for the presence of homologues to known structural RNAs, analogous to the public Pfam search servers.
METHODS
Each family in Rfam is represented by two multiple sequence alignments and a covariance model. The seed alignment contains known representative members of the family, is hand-curated, and is annotated with structural information. The seed alignment is used to build a covariance model using the CMBUILD program from the INFERNAL suite (http://infernal.wustl.edu/) (8). The model is then used to search a nucleotide sequence database using the CMSEARCH program. CMSEARCH reports scores for matches to the model, and a family-specific threshold is chosen such that we believe no false positives fall above the threshold. The matches are then aligned to the model using the CMALIGN program.
The nucleotide database searched is called RFAMSEQ, and is built from a subset of the EMBL nucleotide database (27). RFAMSEQ 1 is based on EMBL release 71. RFAMSEQ includes the ‘finished’ portion of EMBL distributed in the organism specific data files, and excludes the EST, GSS, HTG, HTC, STS and patent sections of the database. Despite these exclusions, RFAMSEQ 1 contains 1 075 317 sequences and over 5.3 billion bases. CM searches are particularly computationally expensive, with a small model (such as tRNA) searching around 200 bases per second on a 600 MHz Compaq ALPHA. A full CM search of RFAMSEQ with one small model would take around 300 cpu days. The search time scales roughly with the cube of the query consensus length, so this quickly becomes entirely infeasible for larger RNAs. We, therefore, employ an initial BLAST search (6) with relaxed search parameters to reduce the search space. All BLAST hits with P-value <10 to a member of the seed alignment are retrieved, a family specific window size added to each end of the matches, and the reduced database subjected to a full CM search. This approach is similar to that employed by the tRNAscan-SE program which uses an heuristic first step followed by full covariance model search (23), but is generally applicable to any ncRNA search. We anticipate that technological and software improvements will in the future allow us to conduct full CM searches to build family alignments.
AVAILABILITY
Rfam is available on the web at http://www.sanger.ac.uk/Software/Rfam/ in the UK, and http://rfam.wustl.edu/ in the US. The database is also available in flatfile format for local installation. To search Rfam locally, the user will also need the INFERNAL software suite, available from http://infernal.wustl.edu/. Table 1 shows a list of families contained in Rfam 1.0. These families annotate over 50 000 ncRNAs in the RFAMSEQ database.
Table 1. Accession numbers and descriptions of the families in Rfam release 1.0. Where data from third party sources are repackaged, or have been used in the construction of seed alignments, the appropriate source is cited.
| Rfam accession | Family description | Third party sources |
|---|---|---|
| RF00001 | 5S ribosomal RNA | 5S ribosomal RNA database (16) |
| RF00002 | 5.8S ribosomal RNA | European LSU rRNA database (11) |
| RF00003 | U1 spliceosomal RNA | The uRNA database (13) |
| RF00004 | U2 spliceosomal RNA | The uRNA database (13) |
| RF00005 | Transfer RNA | |
| RF00006 | Vault RNA | |
| RF00007 | U12 minor spliceosomal RNA | (28) |
| RF00008 | Hammerhead ribozyme | |
| RF00009 | Nuclear RNase P | The Ribonuclease P Database (15) |
| RF00010 | Bacterial RNase P class A | The Ribonuclease P Database (15) |
| RF00011 | Bacterial RNase P class B | The Ribonuclease P Database (15) |
| RF00012 | U3 small nucleolar RNA | The uRNA database (13) |
| RF00013 | 6S/SsrS RNA | |
| RF00014 | DsrA RNA | |
| RF00015 | U4 spliceosomal RNA | The uRNA database (13) |
| RF00016 | U14 small nucleolar RNA | |
| RF00017 | Signal recognition particle RNA | SRPDB (12) |
| RF00018 | CsrB/RsmB RNA | |
| RF00019 | Y RNA (Ro RNP component) | |
| RF00020 | U5 spliceosomal RNA | The uRNA database (13) |
| RF00021 | Spot 42 RNA | |
| RF00022 | GcvB RNA | |
| RF00023 | tmRNA | The tmRNA Website (21) |
| RF00024 | Vertebrate telomerase RNA | (29) |
| RF00025 | Ciliate telomerase RNA | (30) |
WEBSITE FEATURES
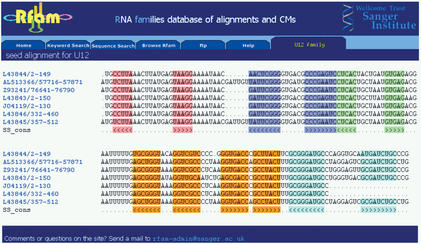
The Rfam websites have been designed to be intuitive to use—users of the Pfam database of protein families will recognise the layout and format of the database. The websites provide the facility to search a DNA sequence against the library of CMs. The user can view annotation on each RNA family, and follow links to other databases and literature references. The multiple sequence alignments on which Rfam is based are available in a number of formats for viewing in a browser or for downloading. Both the seed and full alignments contain secondary structure mark-up to describe the base-paired positions in the member sequences, and the web view provides a colour-encoded representation of these co-varying columns (Fig. 1). In addition the web pages allow the user to quickly determine the species distribution within a family.
Figure 1.
Seed alignment for the U12 spliceosomal RNA family from the UK website. Secondary structure base pairs are encoded in the coloured bases in the alignment, and are marked-up in the SS_cons lines with nested sets of < and > tags. The sequence accessions link to entries in the EMBL database.
FUTURE DIRECTIONS
Rfam is under active development and will increase significantly in size and scope over the next 12 months. Novel ncRNA genes are being discovered at a rapid rate, and we aim to quickly translate such discoveries into useful and searchable RNA families. However, we recognise a number of limitations with our approach. The most obvious of these is the computational cost of using CMs. We predict that technological advances will soon make these searches far more feasible, and will allow full CM genome-wide searches for ncRNAs using Rfam. Until such a time narrowing the search space using BLAST greatly facilitates such searches, though at an inevitable and unknown cost in search sensitivity. In addition, there are RNA families that we cannot model using the alignment- and profile-based approach at present—for example, microRNAs (miRNA precursor secondary structures are only vaguely similar stem-loops) and many small nucleolar RNAs (the consensus of modification guide snoRNAs includes significant inter-molecular base pairing to their target RNAs). Despite such limitations, the Rfam library of alignments and CMs provides a useful tool for genome annotation, as well as a comprehensive resource for RNA family information and multiple sequence alignments.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to William Mifsud for providing annotation for many of the families in Rfam.
REFERENCES
- 1.Eddy S.R. (2002) Computational genomics of noncoding RNA genes. Cell, 109, 137–140. [DOI] [PubMed] [Google Scholar]
- 2.Eddy S.R. (2001) Non-coding RNA genes and the modern RNA world. Nature Rev. Genet., 2, 919–929. [DOI] [PubMed] [Google Scholar]
- 3.Storz G. (2002) An expanding universe of noncoding RNAs. Science, 296, 1260–1263. [DOI] [PubMed] [Google Scholar]
- 4.Pace N.R., Thomas,B.C. and Woese,C.R. (1999) Probing RNA structure, function and history by comparative analysis. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, pp. 113–141.
- 5.Pace N.R. (1997) A molecular view of microbial diversity and the biosphere. Science, 276, 734–740. [DOI] [PubMed] [Google Scholar]
- 6.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eddy S.R. (2002) A memory-efficient dynamic programming algorithm for optimal alignment of a sequence to an RNA secondary structure. BMC Bioinformatics, 3, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eddy S.R. and Durbin,R. (1994) RNA sequence analysis using covariance models. Nucleic Acids Res., 22, 2079–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wuyts J., De Rijk,P., Van de Peer,Y., Winkelmans,T. and De Wachter,R. (2001) The European Large Subunit Ribosomal RNA Database. Nucleic Acids Res., 29, 175–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorodkin J., Knudsen,B., Zwieb,C. and Samuelsson,T. (2001) SRPDB (Signal Recognition Particle Database). Nucleic Acids Res., 29, 169–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zwieb C. (1997) The uRNA database. Nucleic Acids Res., 25, 102–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cannone J.J., Subramanian,S., Schnare,M.N., Collett,J.R., D'Souza,L.M., Du,Y., Feng,B., Lin,N., Madabusi,L.V., ller,K.M., Pande,N., Shang,Z., Yu,N. and Gutell,R.R. (2002) The Comparative RNA Web (CRW) Site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics, 3, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown J.W. (1999) The Ribonuclease P Database. Nucleic Acids Res., 27, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szymanski M., Barciszewska,M.Z., Erdmann,V.A. and Barciszewski,J. (2002) 5S Ribosomal RNA Database. Nucleic Acids Res., 30, 176–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klosterman P.S., Tamura,M., Holbrook,S.R. and Brenner,S.E. (2002) SCOR: a Structural Classification of RNA database. Nucleic Acids Res., 30, 392–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maidak B.L., Cole,J.R., Lilburn,T.G., Parker,C.T.,Jr0a, Saxman,P.R., Farris,R.J., Garrity,G.M., Olsen,G.J., Schmidt,T.M. and Tiedje,J.M. (2001) The RDP-II (Ribosomal Database Project). Nucleic Acids Res., 29, 173–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Batenburg F.H., Gultyaev,A.P. and Pleij,C.W. (2001) PseudoBase: structural information on RNA pseudoknots. Nucleic Acids Res, 29, 194–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wuyts J., Van de Peer,Y., Winkelmans,T. and De Wachter,R. (2002) The European database on small subunit ribosomal RNA. Nucleic Acids Res., 30, 183–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams K.P. (2002) The tmRNA Website: invasion by an intron. Nucleic Acids Res., 30, 179–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knudsen B., Wower,J., Zwieb,C. and Gorodkin,J. (2001) tmRDB (tmRNA database). Nucleic Acids Res., 29, 171–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowe T.M. and Eddy,S.R. (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res., 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laslett D., Canback,B. and Andersson,S. (2002) BRUCE: a program for the detection of transfer–messenger RNA genes in nucleotide sequences. Nucleic Acids Res., 30, 3449–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letunic I., Goodstadt,L., Dickens,N.J., Doerks,T., Schultz,J., Mott,R., Ciccarelli,F., Copley,R.R., Ponting,C.P. and Bork,P. (2002) Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res., 30, 242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falquet L., Pagni,M., Bucher,P., Hulo,N., Sigrist,C.J., Hofmann,K. and Bairoch,A. (2002) The PROSITE database, its status in 2002. Nucleic Acids Res., 30, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoesser G., Baker,W., van den Broek,A., Camon,E., Garcia-Pastor,M., Kanz,C., Kulikova,T., Leinonen,R., Lin,Q., Lombard,V., Lopez,R., Redaschi,N., Stoehr,P., Tuli,M.A., Tzouvara,K. and Vaughan,R. (2002) The EMBL Nucleotide Sequence Database. Nucleic Acids Res., 30, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shukla G.C. and Padgett,R.A. (1999) Conservation of functional features of U6atac and U12 snRNAs between vertebrates and higher plants. RNA, 5, 525–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J.L., Blasco,M.A. and Greider,C.W. (2000) Secondary structure of vertebrate telomerase RNA. Cell, 100, 503–514. [DOI] [PubMed] [Google Scholar]
- 30.McCormick-Graham M. and Romero,D.P. (1995) Ciliate telomerase RNA structural features. Nucleic Acids Res., 23, 1091–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]