Abstract
Sequence conservation in non-coding, upstream regions of orthologous genes from man and mouse is likely to reflect common regulatory DNA sites. Motivated by this assumption we have delineated a catalogue of conserved non-coding sequence blocks and provide the CORG—‘COmparative Regulatory Genomics’—database. The data were computed based on statistically significant local suboptimal alignments of 15 kb regions upstream of the translation start sites of, currently, 10 793 pairs of orthologous genes. The resulting conserved non-coding blocks were annotated with EST matches for easier detection of non-coding mRNA and with hits to known transcription factor binding sites. CORG data are accessible from the ENSEMBL web site via a DAS service as well as a specially developed web service (http://corg.molgen.mpg.de) for query and interactive visualization of the conserved blocks and their annotation.
INTRODUCTION
The COmparative Regulatory Genomics (CORG) database and annotation project aims at providing insights into gene regulation at the level of transcription. Having now several genomes of higher eukaryotes at hand, we are able to study sequence elements on a comparative basis. Comparative sequence analysis has become a powerful tool regarding a variety of problems ranging from gene finding (1,2) to the identification of regulatory elements (3,4). The CORG project systematically applies comparative sequence analysis methods to non-coding, genomic DNA. The working hypothesis underlying the CORG project is that local sequence conservation points to functional importance (5). The CORG project is a resource for the genome-wide annotation of conserved sequence elements in non-coding genomic DNA. We will subsequently call these elements ‘conserved non-coding blocks’ (CNBs). An outline of the derivation of our dataset is given below. Furthermore, an in-depth report on our approach can be found in Dieterich et al. (6).
DETECTION OF CONSERVED NON-CODING BLOCKS
Sequence conservation upstream of two orthologous genes is likely to reflect similar regulatory control of the two genes and, vice versa, many regulatory elements usually cluster in upstream regions. Thus, the CORG project focuses on the detection and collection of conserved blocks from the upstream regions of orthologous genes. We define non-coding conserved blocks as local suboptimal alignments of non-coding genomic DNA of orthologous gene loci. To maximize compatibility with ENSEMBL (7), information on homology of gene loci was retrieved from a compilation of cross-species gene relations in the ENSEMBL compara 7.1 database. The current CORG release 1.0 takes its non-coding upstream sequences from NCBI Human Assembly 29 and the MGSC Mouse Assembly 3.
In order not to bias the analysis by results of earlier computational analysis (e.g. promoter prediction), upstream regions 5′ of the translation start sites of genes are considered. Consequently, these regions have to be allowed a size sufficient to extend at least to the promoter regions of the genes. To determine the extent of this region, promoters described in the EPD database (8) were mapped to the human genome assembly and a histogram of the distances between promoter and start of translation was computed (6). We found that more than 90% of all mapped promoter regions are contained within an interval of 10 kb seen from the start of translation and chose to retrieve and analyze 15 kb of upstream sequence as maximum size. Common mammalian repeats and low complexity regions (e.g. AT-rich) are masked in each sequence pair using Repeatmasker (Smit, A.F.A. and Green, P., http//ftp.genome.washington.edu/RM/RepeatMasker.html).
The next step is the computation of the significant local suboptimal alignments of the sampled pairs of upstream regions. The Waterman–Eggert algorithm (9) is employed to compute these alignments allowing for gaps. Alignments are ranked according to their score. Thus, alignments of the same score have the same rank. Waterman and Vingron (10) showed that the scores of the local suboptimal alignments approximately follow the rank statistic of a Poisson distribution. The parameters to describe this distribution can be derived from simulated alignment data under the same parameters as used in the actual analysis. This facilitates an assessment of the significance of each alignment. Briefly, all suboptimal alignments with a P-value of less than 0.001 were considered significant. The running time of the Waterman–Eggert algorithm is justified by the higher sensitivity aiding the identification of subtle sequence conservation of regulatory sites.
CORG STATISTICS
18 647 homologous gene pairs of man and mouse are defined in the ENSEMBL compara 7.1 database. Significant alignments with a P-value of less than 0.001 were detected in 10 793 gene pairs (58%). This led to the definition of 293 503 CNBs for the two genomes. On average around 8% of each investigated upstream sequence was part of significant conservation. More details on the distributions of scores, lengths and offsets of alignments can be found at http://corg.molgen.mpg.de/stats.html.
ANNOTATION OF CONSERVED NON-CODING BLOCKS
Once the conserved non-coding blocks are defined, they are subject to the annotation process. In this context, matches to ESTs are good indicators for putative exons. Especially, untranslated leader exons in the region upstream of the start of translation can be detected this way. The annotation pipeline computes stringent matches to the GeneNest database (11) containing consensus sequences of assembled EST clusters. In the setting of gene regulation, transcription factor binding sites are another piece of valuable information. The TRANSFAC database (12) is a comprehensive repository of experimentally determined binding site sequences. The annotation pipeline uses TRANSFAC to compute exact matches to known binding site sequences of a minimal length of 8 nucleotides. The co-occurrence of binding sites in the same conserved sequence segment of two or more species underpins their potential role in gene regulation.
THE CORG WEB SERVICE
Internally, CORG data are stored in a relational database containing sequences, alignments, and annotation. There exist two ways of accessing the data via web services. One option is to embed the CORG data in the ENSEMBL ContigView via the distributed annotation system (DAS) (13). Alternatively, one can access the CORG database via the CORG home page (http://corg.molgen.mpg.de). There, the user can query the database by target gene locus. Both accession methods converge at the visualization step.
The distributed annotation system DAS allows for the display of various data sources in a single viewer. Our DAS server (http://tomcat.molgen.mpg.de:8080/das) constitutes such an external data source. Position information of all conserved non-coding blocks and mapped EPD promoters is accessible from this DAS server. Each DAS sequence feature provides a link to the corresponding CORG database entry. New DAS sources can be easily added to the ENSEMBL display. A small tutorial on installing external DAS data sources is available on our web page (http://corg.molgen.mpg.de/DAS_tutorial.htm).
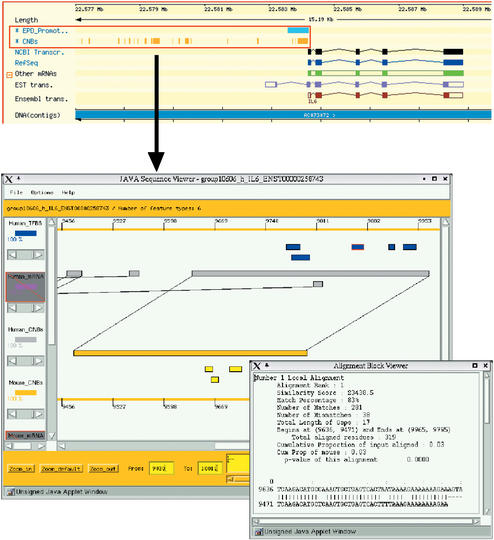
The CORG search page (http://corg.molgen.mpg.de/cgi-bin/cnbsearch.pl) allows querying and accessing the database without using DAS. The user can query the database with standard HUGO gene identifiers or ENSEMBL gene, transcript or protein identifiers. As a search result, a list of all partial matches of the query to database entries is shown. The list serves as a springboard to the visualization step. The combined annotation results are visualized by a JAVA applet (JDK 1.1 standard). This applet runs on all JAVA-compatible web browsers. Detailed information about the conserved non-coding blocks and their arrangement on the corresponding genomes is shown simultaneously. If available, annotation information for putative binding sites of transcription factors and EST matches is displayed on separate tracks. Each individual alignment that defines a CNB can be browsed at the nucleotide level. In addition, web links are assigned to sequence features and allow the user to access external resources like ENSEMBL, GeneNest or TRANSFAC. The applet display is highly adjustable with respect to its appearance and offers an extensive zoom function down to the nucleotide level. Figure 1 gives a complete overview of the CORG user interface.
Figure 1.
Schematic depiction of linking DAS tracks and comparative display in CORG. The upper half of the figure shows a cutout from the ENSEMBL ContigView that is centered on the upstream region of the human IL6 gene. Two DAS tracks showing positions of human EPD promoters and CORG conserved non-coding blocks are highlighted in this context. The user can easily jump to the comparative display that is shown in the lower half of the figure by clicking on any CNB feature. The left bar of the applet window is the legend or key to what is displayed in the window center. Human CNBs are shown in light gray, whereas mouse CNBs are displayed in orange. Putative transcription factor binding sites appear either in blue (human genome) or yellow (mouse genome). Each individual track can be activated or deactivated by clicking on the corresponding legend symbol. Each legend symbol has a lever assigned to it. This lever can be used to adjust the proportion of sequence features displayed for a particular track. Less important sequence features (e.g. low-scoring alignments, low-quality binding sites) will start to disappear first as the lever is moved to lower percentage values. The area below the central display lets you control the zoom level of the display and the exact clipping down to the nucleotide level. Floating menus (not shown) providing links to additional information appear as the mouse is clicked over any feature. Selecting the option ‘Alignment view’ from a floating menu opened the little window on the right. In this window, information on different alignment properties like number, ranking according to score, overall proportion of matches and so on is displayed.
CONCLUSION
The CORG project provides a catalogue of non-coding blocks of DNA sequence conserved between orthologous genes of man and mouse. It is based on a comprehensive, genome-wide computational analysis. Additional annotation information with regard to a potential regulatory function has been included for each catalogue entry. Various earlier studies have shown the advantage of this comparative approach in particular in higher eukaryotes. The CORG database is publicly available via our web service (http://corg.molgen.mpg.de), either for direct query or embedded into the ENSEMBL web site via our DAS server (http://tomcat.molgen.mpg.de:8080/das). We are willing to share the entire data set on a collaborative basis. The CORG project is an ongoing effort meaning that further genome assemblies and annotation steps will be added in the future.
REFERENCES
- 1.Morgenstern B., Rinner,O., Abdeddaim,S., Haase,D., Mayer,K.F., Dress,A.W. and Mewes,H.W. (2002) Exon discovery by genomic sequence alignment. Bioinformatics, 18, 777–787. [DOI] [PubMed] [Google Scholar]
- 2.Wiehe T., Gebauer-Jung,S., Mitchell-Olds,T. and Guigo,R. (2001) SGP-1: prediction and validation of homologous genes based on sequence alignments. Genome Res., 11, 1574–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loots G.G., Ovcharenko,I., Pachter,L., Dubchak,I. and Rubin,E.M. (2002) rVista for comparative sequence-based discovery of functional transcription factor binding. Genome Res., 12, 832–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottgens B., Barton,L.M., Chapman,M.A., Sinclair,A.M., Knudsen,B., Grafham,D., Gilbert,J.G., Rogers,J., Bentley,D.R. and Green,A.R. (2002) Transcriptional regulation of the stem cell leukemia gene (SCL)-comparative analysis of five vertebrate SCL loci. Genome Res., 12, 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardison R.C. Conserved noncoding sequences are reliable guides to regulatory elements. (2000) Trends Genet., 16, 369–372. [DOI] [PubMed] [Google Scholar]
- 6.Dieterich C., Cusack,B., Wang,H., Rateitschak,K., Krause,A. and Vingron,M. (2002) Annotating regulatory DNA based on man-mouse genomic comparison. Bioinformatics, 18 (Suppl 2) 84–90. [DOI] [PubMed] [Google Scholar]
- 7.Hubbard T., Barker,D., Birney,E., Cameron,G., Chen,Y., Clark,L., Cox,T., Cuff,J., Curwen,V., Down,T., Durbin,R., Eyras,E., Gilbert,J., Hammond,M., Huminiecki,L., Kasprzyk,A., Lehvaslaiho,H., Lijnzaad,P., Melsopp,C., Mongin,E., Pettett,R., Pocock,M., Potter,S., Rust,A., Schmidt,E., Searle,S., Slater,G., Smith,J., Spooner,W., Stabenau,A., Stalker,J., Stupka,E., Ureta-Vidal,A., Vastrik,I. and Clamp,M. (2002) The Ensembl genome database project. Nucleic Acids Res., 30, 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Praz V., Perier,R., Bonnard,C. and Bucher,P. (2002) The Eukaryotic Promoter Database, EPD: new entry types and links to gene expression. Nucleic Acids Res., 30, 322–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waterman M.S. and Eggert,M. (1987) A new algorithm for best subsequence alignments with application to tRNA-rRNA comparisons. J. Mol. Biol., 197, 723–728. [DOI] [PubMed] [Google Scholar]
- 10.Waterman M.S. and Vingron,M. (1994) Sequence comparison significance and Poisson approximation. Statistical Science, 9, 367–381. [Google Scholar]
- 11.Haas S.A., Beissbarth,T., Rivals,E., Krause,A. and Vingron,M. (2000) GeneNest: automated generation and visualization of gene indices. Trends Genet., 16, 521–523. [DOI] [PubMed] [Google Scholar]
- 12.Wingender E., Chen,X., Fricke,E., Geffers,R., Hehl,R., Liebich,I., Krull,M., Matys,V., Michael,H., Ohnhauser,R., Pruss,M., Schacherer,F., Thiele,S. and Urbach,S. (2001) The TRANSFAC system on gene expression regulation. Nucleic Acids Res., 29, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dowell R.D., Jokerst,R.M., Day,A., Eddy,S.R. and Stein,L. (2001) The Distributed Annotation System. BMC Bioinformatics, 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]