Abstract
Ribosomal RNA-(rRNA)-targeted oligonucleotide probes are widely used for culture-independent identification of microorganisms in environmental and clinical samples. ProbeBase is a comprehensive database containing more than 700 published rRNA-targeted oligonucleotide probe sequences (status August 2002) with supporting bibliographic and biological annotation that can be accessed through the internet at http://www.probebase.net. Each oligonucleotide probe entry contains information on target organisms, target molecule (small- or large-subunit rRNA) and position, G+C content, predicted melting temperature, molecular weight, necessity of competitor probes, and the reference that originally described the oligonucleotide probe, including a link to the respective abstract at PubMed. In addition, probes successfully used for fluorescence in situ hybridization (FISH) are highlighted and the recommended hybridization conditions are listed. ProbeBase also offers difference alignments for 16S rRNA-targeted probes by using the probe match tool of the ARB software and the latest small-subunit rRNA ARB database (release June 2002). The option to directly submit probe sequences to the probe match tool of the Ribosomal Database Project II (RDP-II) further allows one to extract supplementary information on probe specificities. The two main features of probeBase, ‘search probeBase’ and ‘find probe set’, help researchers to find suitable, published oligonucleotide probes for microorganisms of interest or for rRNA gene sequences submitted by the user. Furthermore, the ‘search target site’ option provides guidance for the development of new FISH probes.
INTRODUCTION
Comparative sequence analysis of ribosomal RNA (rRNA) gene sequences has become the gold standard to infer prokaryotic phylogeny and is widely used in microbial ecology. For example, the application of rRNA-targeted oligonucleotide probes in different hybridization formats such as dot blot (1) and fluorescence in situ hybridization (FISH) (2) allows one to identify uncultured prokaryotes and to quantitatively determine the composition of complex microbial communities (3). Several recent studies also demonstrated the applicability of FISH to routine diagnostic purposes in the clinical laboratory (4–6). In addition, a suite of new techniques circling around rRNA-targeted probes has been developed. rRNA-based phylogenetic DNA microarrays (so-called ‘PhyloChips’) (7–11) consisting of collections of oligonucleotide probes that detect the target microorganisms at multiple taxonomic levels of specificity are now increasingly being developed and applied for diagnostics and environmental microbiology. Furthermore, the combination of FISH and microautoradiography can be used to determine the ecophysiology of microorganisms by visualizing in situ uptake and subsequent incorporation of a radioactively labelled substrate into individual microbial cells (12). As a consequence of the apparent increase in interest in rRNA-targeted oligonucleotide probes during the past years, several hundred ready-to-use domain-, phylum-, genus-, and species-specific probes are already available. However, an overview over published probe sequences can only be obtained by a time-consuming, tedious literature search. Additionally, one has to keep in mind that with the increasing amounts of rRNA sequence data stored in public databases (13,14) (Strunk,O. and Ludwig,W., 1993–2002, ARB—a software environment for sequence data, http://www.arb-home.de) the recognized specificity range for a probe might change. Thus, prior to the application of a rRNA-targeted oligonucleotide probe, researchers are obliged to ascertain that the specificity proposed for this probe in the original publication is still valid. Probe match tools as implemented in the ARB program package (Strunk,O. and Ludwig,W., 1993–2002, ARB—a software environment for sequence data, http://www.arb-home.de) or provided by the Ribosomal Database Project II (RDP-II) (13) offer an option to check for up-to-date specificity of a probe when used in combination with the latest rRNA databases. The pronounced interest of the scientific community in rRNA-targeted oligonucleotide probes is documented by the average 742 user sessions (347 different users) per month recorded for the probe match tool of RDP-II in 2001, making this tool one of the most frequently used software features of the RDP-II website (James R. Cole, personal communication). While oligonucleotide databases for, for example, viral (VirOligo) (15) or human genes (Molecular Probe Data Base) (16) are available, an up-to-date resource for rRNA-targeted oligonucleotide probe sequences for the identification of prokaryotes is currently lacking. In 1996, Alm and coworkers compiled the Oligonucleotide Probe Database (OPD) that listed 96 PCR primers and probes mainly targeting small-subunit (SSU) and large-subunit (LSU) rRNA (17). However, OPD has not been updated since 1997 and is now no longer available through the internet. ProbeBase closes this gap by providing a user-friendly web-interface to search for published oligonucleotide probe sequences and annotated information. Using probeBase, it is possible to search for suitable probes by submitting the name of a target organism or by indicating a certain probe target site. In addition, the ‘find probe set’ tool can be used to rapidly retrieve all published probes perfectly matching rRNA gene query sequences.
ORGANIZATION OF probeBase
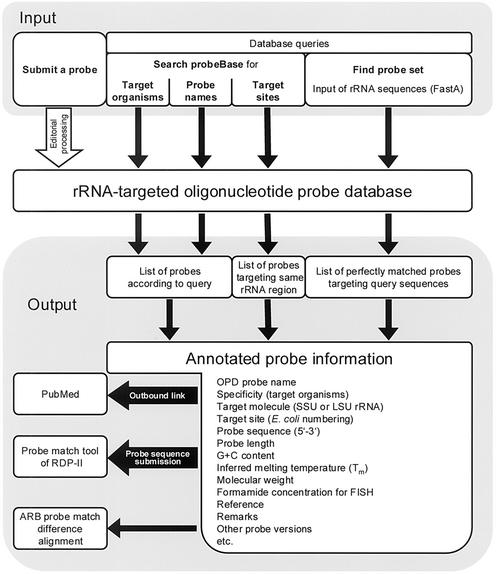
A schematic overview of the structure and organization of probeBase is shown in Figure 1. ProbeBase currently comprises more than 700 published rRNA-targeted oligonucleotide probes (status August 2002). Each probe entry contains information on the probe sequence, target organisms, target molecule (SSU or LSU rRNA), target site, G+C content, melting temperature, molecular weight, and the reference that originally described the oligonucleotide probe. In order to facilitate database searches, each probe entry includes additional hidden information on the taxonomic context of the probe target organisms. If a probe has been successfully applied for FISH, the probe name is highlighted, and the recommended formamide concentration in the hybridization buffer required for specific hybridization is provided. For each probe, probeBase offers direct links to the probe match tool at the RDP-II web site and to the respective reference abstract at PubMed (18). Difference alignments for SSU rRNA-targeted probes are available that were generated using the probe match tool of the ARB software and the ARB database (release June 2002) (Strunk,O. and Ludwig,W., 1993–2002, ARB—a software environment for sequence data, http://www.arb-home.de). Probe entries can also contain supplementary information, such as remarks on the application of the probe or the sequence of a possible competitor probe that has to be used together with the probe to ensure its specificity. In addition, probeBase offers a comprehensive and interactive list ‘Coverage of group-specific probes’ [modified from (19)] showing the coverage of the main prokaryotic lines of descent by general group-specific probes.
Figure 1.
Concept and structure of probeBase.
Search probeBase
ProbeBase can either be searched for probe target organisms, for probe names, or for probe target sites. If probeBase is searched for oligonucleotide probes specific for certain target organisms, it returns a list of all oligonucleotide probes specific for the searched target organisms as well as probes targeting higher taxonomic levels. This list of probes supports researchers in the choice of an appropriate set of nested probes according to the ‘multiple probe concept’ (20). This approach takes advantage of the option to design and apply rRNA-targeted probes for phylogenetic groups at different taxonomic levels (e.g. phylum-, order-, family-, genus-, or species-specific probes). The simultaneous application of a set of hierarchical probes enhances the reliability of the detection of a particular microorganism.
The option to search for a given probe target site assists in the development of new oligonucleotide probes for FISH by providing information on whether a searched target site has previously been found accessible for oligonucleotide probes in other microorganisms. Studies by Fuchs and coworkers have demonstrated that some regions on the 16S and 23S rRNA of Escherichia coli are virtually inaccessible for oligonucleotide probes if used for FISH (21,22). Unfortunately, these results can only be extrapolated to distantly related microorganisms within certain limits. However, if different probes targeting microorganisms affiliated with different evolutionary lineages but sharing the same target site on the respective rRNA molecule have been successfully applied for FISH, it is very likely that the respective target site is generally accessible for oligonucleotide probes.
Find probe set
The ‘find probe set’ tool of probeBase can be used to rapidly retrieve all published probes targeting one or several query rRNA gene sequences without prior comparative sequence analysis. A set of up to 150 sequences, provided by the researcher as rRNA or DNA sense strand sequence in 5′–3′ orientation (FastA format) can be searched simultaneously for the presence of the perfect match target sites of all probes deposited at probeBase. The output is a table sequentially listing (i) each single query sequence with all perfectly matching probes found in probeBase and (ii) each possible probe with all perfectly matching query sequences. Using this probeBase feature researchers will, for example, easily be able to determine a set of already published probes that target the microbial sequences in a certain environmental rRNA gene clone library. This probe set might then be used in subsequent hybridization experiments to confirm the presence of the organisms detected in the rRNA gene clone library in situ and to gain insight into the actual abundance of these microorganisms in the investigated environment.
AVAILABILITY
ProbeBase is maintained and updated by the Microbial Ecology Group staff at the Lehrstuhl für Mikrobiologie of the Technische Universität München, Bavaria, Germany. Free access to probeBase is provided via the world wide web at http://www.probebase.net. Researchers are kindly invited and encouraged to deposit their newly designed probe sequences and supplementary information at probeBase. Submission might either be performed through the probe submission form ‘submit a probe’ accessible at the main page or by Email. For queries concerning probeBase and for alternative probe submission contact probebase@microbial-ecology.net.
CITING probeBase
If you use probeBase as a tool in your published research or if you have deposited your newly designed rRNA-targeted oligonucleotide probes at probeBase, we ask that this paper be cited.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge Wolfgang Ludwig for providing the ARB software and the ARB database to the scientific community—without ARB, research on microbial phylogeny, diversity, and ecology would have been less efficient. We thank James R. Cole from RDP-II for allowing us to directly submit oligonucleotide sequences to the probe match tool of RDP-II and for giving us insight into the probe match user statistics. We also thank Manuela Hartmann and Silvia Weber for their help with database administration, and Klaus Ungerer for his generous support.
REFERENCES
- 1.Raskin L., Poulsen,L.K., Noguera,D.R., Rittmann,B.E. and Stahl,D.A. (1994) Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl. Environ. Microbiol., 60, 1241–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Juretschko S., Loy,A., Lehner,A. and Wagner,M. (2002) The microbial community composition of a nitrifying–denitrifying activated sludge from an industrial sewage treatment plant analyzed by the full-cycle rRNA approach. Syst. Appl. Microbiol., 25, 84–99. [DOI] [PubMed] [Google Scholar]
- 3.Daims H., Ramsing,N.B., Schleifer,K.-H. and Wagner,M. (2001) Cultivation-independent, semiautomatic determination of absolute bacterial cell numbers in environmental samples by fluorescence in situ hybridization. Appl. Environ. Microbiol., 67, 5810–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jansen G.J., Mooibroek,M., Idema,J., Harmsen,H.J., Welling,G.W. and Degener,J.E. (2000) Rapid identification of bacteria in blood cultures by using fluorescently labeled oligonucleotide probes. J. Clin. Microbiol., 38, 814–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu J., Limaye,A.P., Fritsche,T.R., Horn,M., Juretschko,S. and Gautom,R. (2002) Direct detection of Legionellae in respiratory tract specimens using fluorescence in situ hybridization. In Marre,R. (ed.), Legionella. American Society of Microbiology Press, Washington, DC, pp. 221–224.
- 6.Poppert S., Essig,A., Marre,R., Wagner,M. and Horn,M. (2002) Detection and differentiation of chlamydiae by fluorescence in situ hybridization (FISH). Appl. Environ. Microbiol., 68, 4081–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guschin D.Y., Mobarry,B.K., Proudnikov,D., Stahl,D.A., Rittmann,B.E. and Mirzabekov,A.D. (1997) Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl. Environ. Microbiol., 63, 2397–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson K.H., Wilson,W.J., Radosevich,J.L., De Santis,T.Z., Viswanathan,V.S., Kuczmarski,T.A. and Andersen,G.L. (2002) High-density microarray of small-subunit ribosomal DNA probes. Appl. Environ. Microbiol., 68, 2535–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W.T., Mirzabekov,A.D. and Stahl,D.A. (2001) Optimization of an oligonucleotide microchip for microbial identification studies: a non-equilibrium dissociation approach. Environ. Microbiol., 3, 619–629. [DOI] [PubMed] [Google Scholar]
- 10.Wu L., Thompson,D.K., Li,G., Hurt,R.A., Tiedje,J.M. and Zhou,J. (2001) Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol., 67, 5780–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loy A., Lehner,A., Lee,N., Adamczyk,J., Meier,H., Ernst,J., Schleifer,K.-H. and Wagner,M. (2002) Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol., 68, 5064–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee N., Nielsen,P.H., Andreasen,K.H., Juretschko,S., Nielsen,J.L., Schleifer,K.-H. and Wagner,M. (1999) Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol., 65, 1289–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maidak B.L., Cole,J.R., Lilburn,T.G., Parker,C.T., Jr., Saxman,P.R., Farris,R.J., Garrity,G.M., Olsen,G.J., Schmidt,T.M. and Tiedje,J.M. (2001) The RDP-II (Ribosomal Database Project). Nucleic Acids Res., 29, 173–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wuyts J., Van de Peer,Y., Winkelmans,T. and De Wachter,R. (2002) The European database on small subunit ribosomal RNA. Nucleic Acids Res., 30, 183–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Onodera K. and Melcher,U. (2002) VirOligo: a database of virus-specific oligonucleotides. Nucleic Acids Res., 30, 203–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campi M.G., Romano,P., Milanesi,L., Marra,D., Manniello,M.A., Iannotta,B., Rondanina,G., Grasso,E., Ruzzon,T. and Santi,L. (1998) Molecular Probe Data Base (MPDB). Nucleic Acids Res., 26, 145–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alm E.W., Oerther,D.B., Larsen,N., Stahl,D.A. and Raskin,L. (1996) The oligonucleotide probe database. Appl. Environ. Microbiol., 62, 3557–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheeler D.L., Church,D.M., Lash,A.E., Leipe,D.D., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Tatusova,T.A., Wagner,L. and Rapp,B.A. (2002) Database resources of the National Center for Biotechnology Information: 2002 update. Nucleic Acids Res., 30, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loy A., Daims,H. and Wagner,M. (2002) Activated sludge-Molecular techniques for determining community composition. In Bitton,G. (ed.), The Encyclopedia of Environmental Microbiology. John Wiley & Sons, Inc., New York, pp. 26–43.
- 20.Behr T., Koob,C., Schedl,M., Mehlen,A., Meier,H., Knopp,D., Frahm,E., Obst,U., Schleifer,K., Niessner,R. and Ludwig,W. (2000) A nested array of rRNA targeted probes for the detection and identification of enterococci by reverse hybridization. Syst. Appl. Microbiol., 23, 563–572. [DOI] [PubMed] [Google Scholar]
- 21.Fuchs B.M., Syutsubo,K., Ludwig,W. and Amann,R. (2001) In situ accessibility of Escherichia coli 23S rRNA to fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol., 67, 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuchs B.M., Wallner,G., Beisker,W., Schwippl,I., Ludwig,W. and Amann,R. (1998) Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol., 64, 4973–4982. [DOI] [PMC free article] [PubMed] [Google Scholar]