Abstract
The Medaka Expression Pattern Database (MEPD) stores and integrates information of gene expression during embryonic development of the small freshwater fish Medaka (Oryzias latipes). Expression patterns of genes identified by ESTs are documented by images and by descriptions through parameters such as staining intensity, category and comments and through a comprehensive, hierarchically organized dictionary of anatomical terms. Sequences of the ESTs are available and searchable through BLAST. ESTs in the database are clustered upon entry and have been blasted against public data-bases. The BLAST results are updated regularly, stored within the database and searchable. The MEPD is a project within the Medaka Genome Initiative (MGI) and entries will be interconnected to integrated genomic map databases. MEPD is accessible through the WWW at http://medaka.dsp.jst.go.jp/MEPD.
INTRODUCTION
The knowledge of how a gene is expressed in time and space is the first clue to its function. Expression patterns can be determined in an easy, straightforward approach through in situ hybridization. Large-scale in situ hybridization screens have been carried out for the major model organisms in developmental biology like Caenorhabditis elegans (1), Drosophila (2), Ascidia (3), Zebrafish (4), Xenopus (5) and mouse (6) and expression data is stored in databases and accessible through web interfaces. Medaka has developed into a standard developmental model organism over the past years (7). In Medaka in situ screens have been carried out (8,9), but a public database has not been established yet. Large-scale screens are in process in the Medaka scientific community as well as within the Medaka Genome Initiative (MGI) (http://www.dsp.jst.go.jp/MGI) increasing the number of known expression patterns tremendously in the near future. Currently the database contains data from Wittbrodt's group, which will continue submitting data. The database is open for submissions from other groups through web interfaces and access is free to the scientific community. Clones can be ordered from J. Wittbrodt.
DATA
General
To date, the Medaka Expression Pattern Database (MEPD) contains information on 711 clones of a Medaka cDNA plasmid library [CAB-strain, stage 24 (16 somite stage)], 573 sequences in 473 clusters and 1482 in situ pattern images. The clone ID (accession number) consists of the library name (631) and a clone number, which corresponds to the plate number and the coordinates of a clone in a 384 well plate (for example: 631-134-02-C). A servlet displaying all available information concerning a single clone can be accessed as follows: http://medaka.dsp.jst.go.jp:10080/servlet/ShowClone?cloneID=631-13-402-C. This link can be used to access a MEPD entry directly from other databases.
Two other groups of the MGI will submit their data to this database in the near future using different libraries and we encourage any group to submit their data to MEPD.
Expression patterns
Expression patterns are specified at three different embryonic stages (st19: two somite stage, st24: 16 somite stage, st32: somite completion stage) through pictures and a pattern description. Pictures and descriptions complement each other. Stained structures which are not visible in detail in the pictures are further specified in the description.
Expression patterns have been described through text comments, signal intensity (weak, medium, strong), category (specific, differential and homogenous) and through a dictionary of anatomical structures developed for this database (Figs 1 and 2).
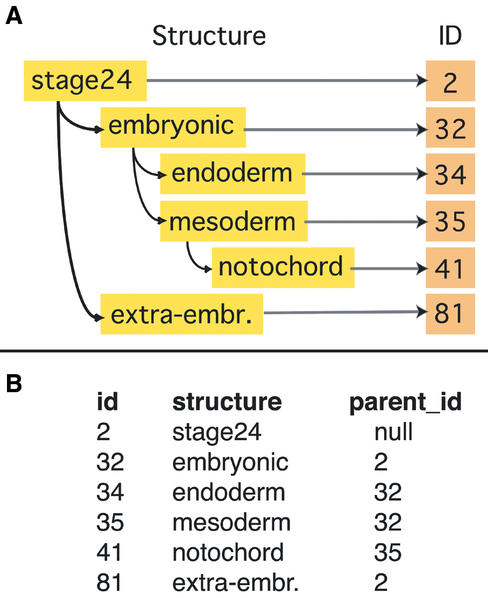
Figure 1.
(A) Expression patterns are described by comments, intensity, category and a hierarchically organized dictionary of anatomical terms. (B) Representation of this hierarchical dictionary in a database table with id, structure and parent_id, which is a key to its higher level id.
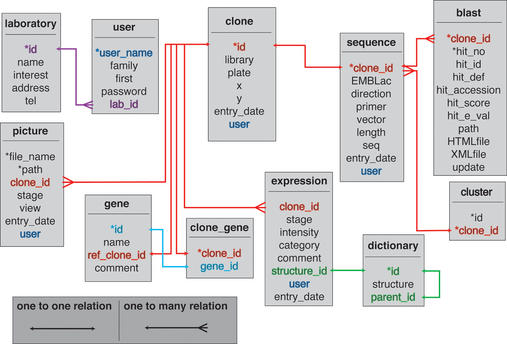
Figure 2.
Entity Relation Diagram of MEPD. The foreign keys to the table user have been omitted to ease an overview, but corresponding column names are highlighted in blue.
This dictionary has been structured in accordance to the mouse dictionary (10) in a hierarchical tree-like organization. In the database table, it has been realized as rows with an identity, a structure name and a key to its parent structure (Fig. 1). This hierarchical organization facilitates a fast database search and enables flexible updating and extension of the dictionary. It allows the refinement of a pattern description from crude to detailed when describing an expression pattern during data submission.
Forty-four (10.76%) clones or 13.77% of the genes were categorized as homogenous, 150 (36.67%) clones or 33.6% of the genes as differential and 215 (52.57%) clones or 52.63% of the genes as specific. Whole mount in situ detection has been performed using a robot (‘InsituPro’ from intavis) with a standard protocol (11).
Sequences
Clones which were used to transcribe the in situ probes are described by their sequence, the sequencing primer, the vector used and the library they come from (Fig. 2). All sequences entered so far have been sequenced from the 5′ end. Sequences have been cleaned from vector contamination, submitted to the EMBL nucleotide database and are linked to it over their accession number. The sequences are blasted against public databases at NCBI using blastn and the 10 closest BLAST results are stored within the database (Fig. 2). The oldest BLAST results are updated by an automated script on a daily basis. Sequences in the database have been clustered using the blastclust program of the standalone BLAST package (12) and cluster information is stored in a database table (Fig. 2).
Pictures
Pictures are stored on the file system of the database server in JPEG format (∼220 kb each, 330 MB in total). Path, filename, view and embryonic stage are stored within the database (Fig. 2).
Other
User and group information is kept in database tables (Fig. 2) to keep track of data submission or eventual access rights.
A non redundant list of genes is stored in the table gene (Fig. 2). One representative clone for each cluster has been entered into this table. A gene name has been assigned to genes showing significant homology to known genes.
DATA ACCESS
Implementation of MEPD
Data is stored in a relational database (IBM DB2) on an IBM RS6000 under AIX in Kyoto. Access is enabled through Java Servlets running on a WebSphere administration server. The Java Servlets receive a client request, create and send SQL-commands to the database server, use the result sets to create a HTML response and send it back to the client. The MEPD is accessible on the World Wide Web at http://medaka.dsp.jst.go.jp/MEPD.
Java Servlets are fast and flexible as they run within the server and do not need to start child processes like CGI programs. They inherit the strong safety of the Java language. Object oriented programming allows the development of reusable code. Using the standard languages SQL and Java facilitates the portability of the software developed here to other platforms. The structure of the SQL tables of the MEPD is shown in Figure 2.
Query interfaces
ESTs can be searched by complete or partial clone identity, for example, the cDNA library or plate number. A text string can be searched in the BLAST results definition in order to find homologous genes or certain species. The search can be restricted to significant homologies only, or ‘new’ genes (with low homology to any known gene) can be selected. A text string can also be searched in the gene annotation.
One can search for the expression pattern by expression intensity, category and anatomical terms of stained structures. The hierarchical implementation of the dictionary allows the inclusion of substructures in the search. By using an alphabetical view of the dictionary one can search for a certain structure without its substructures.
Using a combined search the search parameters described above can be combined.
Finally one can do a sequence homology search by submitting a query sequence (nucleotide or protein) which is blasted against sequences in the database using the blastn, tblastn or tblastx programs of the standalone BLAST package (12). Cluster information can be accessed here.
The result is displayed as a list of clones with abbreviated information, which can be completed through further links.
Data submission
Web interfaces have been developed which allow the online submission of pictures, sequences and pattern descriptions clone by clone. Sequences and images can also be entered through bunch submissions. For either, please contact the authors.
FUTURE PERSPECTIVE
As not only submission by Wittbrodt's group will continue in the future, but also other groups will join in submitting data, MEPD will grow steadily. To guarantee comparability of data, future submissions will be stored in the same standardized way described here. MEPD, therefore, can serve as a resource of Medaka expression information for the scientific community, for example as a marker collection to analyze Medaka mutants.
Within the MGI, an initiative of Medaka researchers, the ESTs of the MEPD are currently being and will be mapped both physically on a radiation hybrid panel and on a Bac-contig map, as well as genetically on a meiotic map. Links to and from these mapping databases will be established. Modules displaying combined data sets of these databases will be developed, which will allow the linkage between expression data and genomic location.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Bjoern Kindler for helping us solve Java Servlet- and SQL-programming problems. We appreciate the contribution of Franck Bourrat in the generation of the Medaka anatomical dictionary. We would like to thank Hiroshi Suwa for his technical help and Hans Doebbeling for enabling us to use the computational facilities of EMBL in Heidelberg.
REFERENCES
- 1.Martinelli S.D., Brown,C.G. and Durbin,R. (1997) Gene expression and development databases for C. elegans. Semin. Cell Dev. Biol., 8, 459–467. [DOI] [PubMed] [Google Scholar]
- 2.Janning W. (1997) FlyView, a Drosophila image database, and other Drosophila databases. Semin. Cell Dev. Biol., 8, 469–475. [DOI] [PubMed] [Google Scholar]
- 3.Kawashima T., Kawashimal,S., Kohara,Y., Kanehisa,M. and Makabe,K.W. (2002) Update of MAGEST: Maboya gene expression patterns and sequence tags. Nucleic Acids Res., 30, 119–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kudoh T., Tsang,M., Hukriede,N.A., Chen,X., Dedekian,M., Clarke,C.J., Kiang,A., Schultz,S., Epstein,J.A., Toyama,R. and Dawid,I.B. (2001) A gene expression screen in Zebrafish embryogenesis. Genome Res., 11, 1979–1987. [DOI] [PubMed] [Google Scholar]
- 5.Pollet N., Schmidt,H.A., Gawantka,V., Vingron,M. and Niehrs,C. (2000) Axeldb: a Xenopus laevis database focusing on gene expression. Nucleic Acids Res., 28, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ringwald M., Eppig,J.T., Begley,D.A., Corradi,J.P., McCright,I.J., Hayamizu,T.F., Hill,D.P., Kadin,J.A. and Richardson,J.E. (2001) The Mouse Gene Expression Database (GXD). Nucleic Acids Res., 29, 98–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wittbrodt J., Shima,A. and Schartl,M. (2002) Medaka—a model organism from the far East. Nature Rev. Genet., 3, 53–64. [DOI] [PubMed] [Google Scholar]
- 8.Henrich T. and Wittbrodt,J. (2002) An in situ hybridization screen for the rapid isolation of differentially expressed genes. Dev. Genes Evol., 210, 28–33. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen V., Joly,J. and Bourrat,F. (2001) An in situ screen for genes controlling cell proliferation in the optic tectum of the medaka (Oryzias latipes). Mech. Dev., 107, 55–67. [DOI] [PubMed] [Google Scholar]
- 10.Bard J.L., Kaufman,M.H., Dubreuil,C., Brune,R.M., Burger,A., Baldock,R.A. and Davidson,D.R. (1998) An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature. Mech Dev., 74, 111–120. [DOI] [PubMed] [Google Scholar]
- 11.Hauptmann G. and Gerster,T. (1994) Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet., 10, 266. [DOI] [PubMed] [Google Scholar]
- 12.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]