Abstract
The Nuclear Protein Database (NPD) is a curated database that contains information on more than 1300 vertebrate proteins that are thought, or are known, to localise to the cell nucleus. Each entry is annotated with information on predicted protein size and isoelectric point, as well as any repeats, motifs or domains within the protein sequence. In addition, information on the sub-nuclear localisation of each protein is provided and the biological and molecular functions are described using Gene Ontology (GO) terms. The database is searchable by keyword, protein name, sub-nuclear compartment and protein domain/motif. Links to other databases are provided (e.g. Entrez, SWISS-PROT, OMIM, PubMed, PubMed Central). Thus, NPD provides a gateway through which the nuclear proteome may be explored. The database can be accessed at http://npd.hgu.mrc.ac.uk and is updated monthly.
INTRODUCTION
Determining the cellular localisation of proteins is important for understanding genome regulation and function, as well as providing important clues as to the molecular function of novel proteins (1). The Nuclear Protein Database (NPD), which began as a repository for data on novel nuclear proteins isolated by a mammalian gene-trap screen (2), provides an overview of the diversity of many subnuclear compartments. As well as gene-trap proteins, more than 1300 vertebrate nuclear proteins reported in the literature have also been archived in NPD. Thus, NPD provides much needed annotation for the nuclear proteome.
DATABASE CONTENT AND STRUCTURE
NPD is a MySQL database that is queried using PHP. The database includes information on gene sequence and chromosomal localisation, and information on protein sequence such as predicted protein size, isoelectric point, as well as any repeats, motifs or domains within the protein sequence. In general only one isoform of the protein is given (usually the largest). Orthologous proteins from different species are also stored under each entry. Homologous proteins are also recorded as ‘related’ proteins. The database contains no information on protein isoforms generated by alternative splicing. Where appropriate, links to other databases are provided [e.g. Entrez (3), Swiss-Prot (4), OMIM (5), PubMed (3), PubMed Central (6)]. Biological and molecular functions of the proteins are described using Gene Ontology (GO) terms (7). An overview of the structure of NPD is show in Figure 1. Additional material available on the website includes descriptions of subnuclear compartments, statistics and links to other relevant databases and resources.
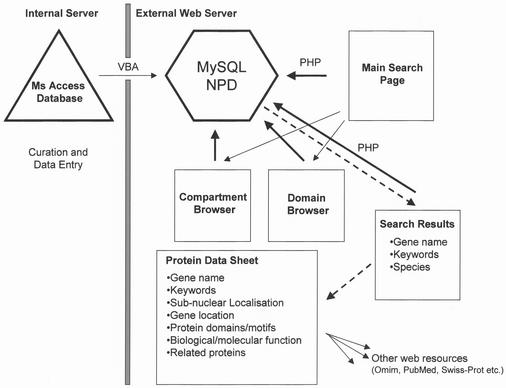
Figure 1.
Overview of the Nuclear Protein Database (NPD). Data is entered into a dedicated MS Access database (triangle) and once a month the Access data tables are converted to MySQL tables using MS Visual Basic (VBA). The SQL tables are then copied to the external MySQL server (hexagon) to be queried using PHP (thick arrows). NPD can be searched using Boolean operators via the web using the main search page. Alternately, the database may be queried by compartment (Compartment Browser) or protein domain (Domain Browser). Search results (dashed arrows) are returned as a meta-list of matching gene entries. The individual entries are retrieved by clicking on the main gene name, which queries the MySQL server once again returning a protein data sheet (PDS) containing archived and external database information. All result and PDS web pages are generated dynamically using PHP. Web pages are represented by squares.
DATABASE ACCESS
The database is available on the www at http://npd.hgu.mrc.ac.uk.
The database is searchable by gene or protein name, gene localisation, species, protein domain, nuclear compartment, external database unique identifiers, GO term, experiment and keyword. Logical queries can be built using Boolean operators. Searches can be restricted to nuclear compartments and protein domains. Alternatively, the database can be browsed using an alpha list of protein domains (domain browser), which includes links to Pfam (8), InterPro (9) and SMART (10). The database may also be searched by sub-nuclear compartment using the compartment browser (Fig. 2), which provides illustrations and descriptions of the various sub-nuclear compartments.
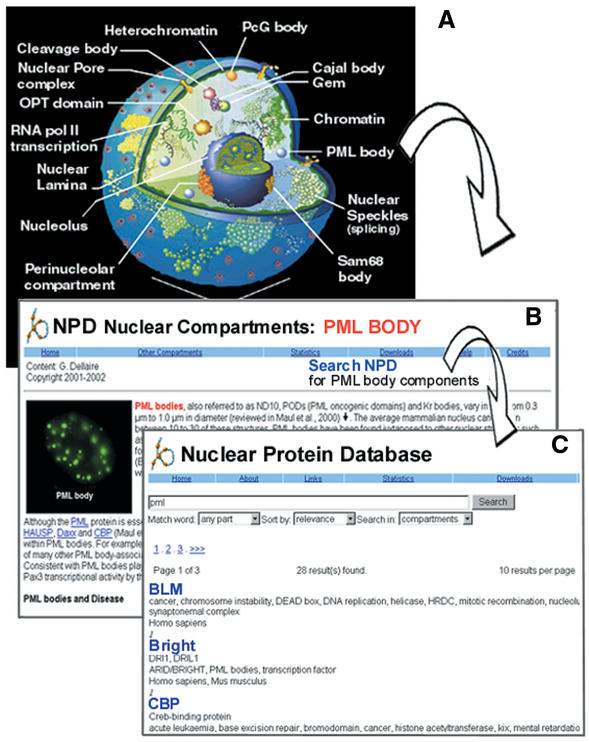
Figure 2.
The NPD Compartment Browser. The compartment browser (A) provides an overview of each of the principal sub-nuclear compartments. Upon selecting a compartment (i.e. PML) an overview page (B) is generated with an image of that compartment and a short abstract describing its structure, function and relevance to human disease. Links to external Internet resources are also provided. In addition, the NPD can be searched for proteins associated with a given domain (C) from the overview page (i.e. BLM, Bright and CBP are returned for PML bodies). Nucleus illustration reproduced by permission of Dr David L. Spector, Cold Spring Harbor Laboratory.
During a query the results are returned as 10 entries per page listing the main gene name, alternate name(s), species and keywords. Search output can be organised by either relevance or alphabetically by gene name using drop-down boxes. The user can navigate the result pages by clicking individual page numbers and can access an entry by clicking on the main gene name. Additional search terms can be added to refine a search using Boolean operators (e.g. kinase AND cytokinesis). In addition, drop-down boxes provide the ability to limit searches to a particular sub-nuclear compartment. Once an entry is chosen a protein data sheet (PDS) for that protein is displayed containing various information on that protein as discussed above. Relevant external database links are also displayed from the PDS allowing the user to retrieve further information (e.g. protein sequence, PubMed entries etc.).
FUTURE PERSPECTIVES
We are continuing to expand the NPD database to include links to additional databases and on-line resources, as they become available. The NPD is a curated database and all information is supported by links to published material. NPD is also updated approximately once a month, thus ensuring the timely reporting of data regarding nuclear proteins. Future planned improvements will include web-based data submission for NPD users, regular GO updates and integration with both Swiss-Prot (4) and the SRS system of the European Bioinformatics Institute (EBI) (http://srs.ebi.ac.uk). Database files, for the extraction of Accession numbers required for BLAST searches, are available upon request. In future, these files and a BLAST facility will be available directly from the web site.
As a bioinformatics tool, we have used NPD to determine a number of important correlations between domain structure and primary sequence characteristics, and the subnuclear compartmentalisation of nuclear proteins (1). For example, we have noted that the proteins that concentrate in the splicing speckle compartments of the nucleus have very high pI values (>11), and that proteins located at the nuclear periphery or in PML/ND10 bodies are surprisingly large and acidic in nature (1). Using these observations, we hope to develop a set of algorithms for structural genomics that could be used for the prediction of sub-nuclear localisation from primary protein sequence, and the identification of novel protein domains (11,12). NPD provides a resource for researchers as well as a gateway for students to explore the complexity of the mammalian nucleus.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to acknowledge the help of MRC HGU computing services and Dr Heidi Sutherland for comments and data entry during the development of NPD. We especially thank all colleagues who have contributed to the development and information content of NPD. The NPD was made possible by funding from the Medical Research Council (UK) and the James S. McDonnell Foundation. G.D. was supported by a fellowship from the Canadian Institutes of Health Research (CIHR) and W.A.B. is a Centennial Fellow of the James S. McDonnell Foundation.
REFERENCES
- 1.Bickmore W.A. and Sutherland,H.G.E. (2002) Addressing protein localisation in the nucleus. EMBO J., 21, 1248–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sutherland H.G.E., Mumford,G.K., Newton,K., Ford,L.V., Farrall,R., Dellaire,G., Cáceres,J.F. and Bickmore,W.A. (2001) Large-scale identification of mammalian proteins localized to nuclear sub-compartments. Hum. Mol. Genet., 10, 1995–2011. [DOI] [PubMed] [Google Scholar]
- 3.Wheeler D.L., Church,D.M., Lash,A.E., Leipe,D.D., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Tatusova,T.A., Wagner,L. and Rapp,B.A. (2001) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 29, 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamosh A., Scott,A.F., Amberger,J., Bocchini,C., Valle,D. and McKusick,V.A. (2002) Online Mendelian Inheritance in Man (OMIM), a knowledge-base of human genes and genetic disorders. Nucleic Acids Res., 30, 52–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts R.J. (2001) PubMed Central: The GenBank of the published literature. Proc. Natl Acad. Sci. USA., 98, 381–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Gene Ontology Consortium. (2000) Gene Ontology: tool for the unification of biology. Nature Genet., 25, 25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Apweiler R., Attwood,T.K., Bairoch,A., Bateman,A., Birney,E., Biswas,M., Bucher,P., Cerutti,L., Corpet,F., Croning,M.D. et al. (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res., 29, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Letunic I., Goodstadt,L., Dickens,N.J., Doerks,T., Schultz,J., Mott,R., Ciccarelli,F., Copley,R.R., Ponting,C.P. and Bork,P. (2002) Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res., 30, 242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisenhaber F. and Bork,P. (1998) Wanted: subcellular localization of proteins based on sequence. Trends Cell Biol., 8, 169–170. [DOI] [PubMed] [Google Scholar]
- 12.Ponting C.P. (2001) Issues in predicting protein function from sequence. Brief. Bioinform., 2, 19–29. [DOI] [PubMed] [Google Scholar]