Abstract
The EMBL Nucleotide Sequence Database (http://www.ebi.ac.uk/embl/) incorporates, organizes and distributes nucleotide sequences from all available public sources. The database is located and maintained at the European Bioinformatics Institute (EBI) near Cambridge, UK. In an international collaboration with DDBJ (Japan) and GenBank (USA), data are exchanged amongst the collaborating databases on a daily basis to achieve optimal synchronization. Webin is the preferred web-based submission system for individual submitters, while automatic procedures allow incorporation of sequence data from large-scale genome sequencing centres and from the European Patent Office (EPO). Database releases are produced quarterly. Network services allow free access to the most up-to-date data collection via FTP, Email and World Wide Web interfaces. EBI's Sequence Retrieval System (SRS) integrates and links the main nucleotide and protein databases plus many other specialized molecular biology databases. For sequence similarity searching, a variety of tools (e.g. Fasta, BLAST) are available which allow external users to compare their own sequences against the latest data in the EMBL Nucleotide Sequence Database and SWISS-PROT. All resources can be accessed via the EBI home page at http://www.ebi.ac.uk.
INTRODUCTION
The European Bioinformatics Institute (EBI), an Outstation of the European Molecular Biology Laboratory (EMBL) in Heidelberg (Germany), is located on the Wellcome Trust Genome Campus near Cambridge (UK), together with the Sanger Institute and the Human Genome Mapping Resource Centre (HGMP-RC). Building, maintaining and providing biological databases and information services to support data deposition and data exploitation are the main missions of the Service Programme of the EBI (1). Databases operated at the EBI include the EMBL Nucleotide Sequence Database (aka as EMBL-Bank), protein databases SWISS-PROT & TrEMBL (2), InterPro (3), the Macromolecular Structure Database (E-MSD) (4), ArrayExpress for gene expression data (5), ENSEMBL (6) for automatic genome annotation plus several other databases many of which are produced in collaboration with external groups.
In Europe, the vast majority of all nucleotide sequence data generated and published are collected, organized and distributed by the EMBL Nucleotide Sequence Database, the European member of the tri-partide International Nucleotide Sequence Database Collaboration DDBJ/EMBL/GenBank, managing sequence data worldwide since 1982. Main sources of data are large-scale genome sequencing projects, direct submissions by individual scientists plus sequence data extracted from BIOTECH patent applications to the European Patent Office. To achieve optimal synchronization, all new and updated database records are exchanged on a dialy basis between EMBL, DDBJ (Japan) (7) and GenBank (USA) (8).
Within a 12 month period the database size has increased from about 12.9 million entries comprising 13.8 Gigabases (Release 68, September 2001) to 18.3 million entries and over 23 Gigabases (Release 72, September 2002). The database growth has been a direct consequence of ongoing collaborations with sequencing projects like the Mouse Genome Sequencing Consortium (MGSC), the International Anopheles Genome Project and a growing number of other genome sequencing groups producing large quantities of new sequence data. During the same period the number of organisms represented in the database has risen by ∼25% to over 100 000 species.
Major new developments during 2002 (described in detail below) include the creation of the CON(struct) or CON(tig) database division, the EMBL Sequence Version Archive, the Whole Genome Shotgun (WGS) Sequences data collection and the Third Party Annotation (TPA) dataset. For a more detailed description of EMBL Nucleotide Sequence Database activites, please see URL http://www.ebi.ac.uk/embl/.
SUBMISSIONS TO EMBL-BANK
Why is it essential to submit new sequences and annotations?
Just try to imagine, where molecular biology and genome research would be today without free access to the comprehensive collection of nucleotide sequences and biological annotations provided by EMBL-Bank in collaboration with DDBJ and GenBank for nearly two decades now. The up-to-date repository of primary nucleotide sequences is an essential requirement for further computational analysis and genome research and todays' molecular biologists depend on free access to all nucleotide sequence data available worldwide. Discovery of novel genes, identification of homologous genes, analysis of alternative splicing and detection of polymorphisms are only some of the uses of the database in the context of biomedical research, and this will only increase as large-scale sequencing efforts keep on depositing more high-throughput sequence data (HTG) and as more finished genomes are being added to the database.
How to submit new sequences and annotations?
Only the basic submission procedures are described in this context, for detailed information on sequence submissions to the EBI (including genome data, alignments, bulk, updates, vector screening etc please see URL http://www.ebi.ac.uk/embl/Submission/.
WEBIN
‘Webin’ is EMBL's preferred submission system for nucleotide sequences and biological annotation information. Webin is designed to allow fast submission of single, multiple or very large numbers of sequences (bulk submissions) and is available at URL: http://www.ebi.ac.uk/embl/Submission/webin.html.
Genome project submissions
EBI staff work closely with sequencing centers to ensure timely incorporation of new data into EMBL-Bank for public release. Database entries produced at the research site are deposited and updated directly by the genome project submitters using FTP or Email. Groups producing large volumes of genome sequence data over an extended period of time are encouraged to submit to an EMBL submission account and are advised to contact the database at datasubs@ebi.ac.uk.
Alignment submissions
‘Webin-Align’ is a dedicated web-based submission tool for submission of multiple sequence alignments in all common alignment formats. ‘Webin-Align’ is available at http://www.ebi.ac.uk/embl/Submission/align_top.html. EMBL-Align (9) is a public dataset of multiple sequence alignments and can be queried from the EBI-SRS server.
ACCESS TO EMBL'S NUCLEOTIDE SEQUENCE DATA
The up-to-date nucleotide sequence data collection is available from EBI's network services. Access is also granted via Email using the netserver or interactively via the WWW, where the main service comprises the SRS server. EMBL datasets are freely available from EBI's FTP-server at ftp://ftp.ebi.ac.uk/pub/databases/embl/. For more information see URL http://www.ebi.ac.uk/embl/Access/ For a complete list of EMBL-Bank internet-based resources see Table 1.
Table 1. EMBL-Bank internet-based resources including detailed information on submissions, data access, genome data as well as database searching and analysis tools.
| Title | URL |
|---|---|
| General | |
| EMBL-EBI Home Page | www.ebi.ac.uk/ |
| EMBL Nucleotide Sequence Database | www.ebi.ac.uk/embl/ |
| Documentation | |
| EMBL Database Documentation Page | www.ebi.ac.uk/embl/Documentation/ |
| Database User manual | www.ebi.ac.uk/embl/Documentation/User_manual/usrman.html |
| Database Release Notes | www.ebi.ac.uk/embl/Documentation/Release_notes/current/relnotes.html |
| The DDBJ/EMBL/GenBank Feature Table Document | www.ebi.ac.uk/embl/Documentation/FT_definitions/feature_table.html |
| Taxonomy Database | www3.ncbi.nlm.nih.gov/Taxonomy/tax.html |
| Example Database Entry | www.ebi.ac.uk/cgi-bin/emblfetch?x64011 |
| WEBALIGN: Sequence Alignment Submissions | www.ebi.ac.uk/embl/Submission/alignment.html |
| WebFeat: feature table keys/qualifiers definitions | www3.ebi.ac.uk/Services/WebFeat/ |
| Annotation Examples: EMBL entry examples. | www3.ebi.ac.uk/Services/Standards/web/ |
| DE line Standards: guidelines for entry definitions | www.ebi.ac.uk/embl/Documentation/de_line_standards.html |
| Sites maintaining daily updated copies of EMBL | www.ebi.ac.uk/embl/Access/other_sites.html |
| Submissions | |
| Submission of Nucleotide Sequence Data | www.ebi.ac.uk/embl/Submission/ |
| Information for Submitters Document | www.ebi.ac.uk/embl/Documentation/information_for_submitters.html |
| Vector Scanning prior to submission | www2.ebi.ac.uk/blastall/vectors.html |
| WEBIN: web-based sequence submission system | www.ebi.ac.uk/embl/Submission/ |
| SEQUIN: stand-alone sequence submission tool | www3.ebi.ac.uk/Services/Sequin/ |
| Genome Project Submission Account guidelines | www3.ebi.ac.uk/Services/GenomeSubm/ |
| WEBUP: sequence update form | www3.ebi.ac.uk/Services/webin/update/update.html |
| Access | |
| Access to servers, query tools and data archives | www.ebi.ac.uk/embl/Access/ |
| Sequence Retrieval Service (SRS) | http://srs.ebi.ac.uk/ |
| FTP server | ftp://ftp.ebi.ac.uk/pub/databases/embl/ |
| Current Database Release | ftp://ftp.ebi.ac.uk/pub/databases/embl/release/ |
| Sequence Tagged Sites (STS) resources | www.ebi.ac.uk/embl/Access/sts.html |
| Expressed Sequence Tag (EST) resources | www.ebi.ac.uk/embl/Access/est.html |
| Third Party Annotation Database (TPA) | ftp://ftp.ebi.ac.uk/pub/databases/embl/tpa/ |
| Sequences from the patent literature | ftp://ftp.ebi.ac.uk/pub/databases/embl/patent/ |
| Genome Data | |
| Completed Genomes Web Server | www.ebi.ac.uk/genomes/ |
| Genome FTP server | ftp://ftp.ebi.ac.uk/pub/databases/embl/genomes/ |
| Whole Genome Shotgun Dataset | ftp://ftp.ebi.ac.uk/pub/databases/embl/wgs/ |
| EnsEMBL: automated analysis of genome data | http://ensembl.ebi.ac.uk/ |
| Genome MOT: status of genome projects | www.ebi.ac.uk/Databases/Genome_MOT/genome_mot.html |
| Database searching, browsing and analysis tools | |
| Access to searching, browsing and analysis tools | www.ebi.ac.uk/Tools/ |
Completed genome sequences
New genomes included in the database over the last 12 months include Plasmodium falciparum, Schizosaccharomyces pombe, Streptomyces coelicolor (model actinomyce) plus many others. Direct access to several hundreds of completed genome sequences is available via EBI's WWW Genomes server at http://www.ebi.ac.uk/genomes/.
Unfinished genome sequences
Unfinished genomic data is incorporated into the EMBL Database high-throughput genome division (HTG) or the whole genome shotgun (WGS) dataset. WGS data for Oryza sativa, Anopheles gambiae, Mus musculus, Bacillus anthracis, Takifugu rubripes and other organisms are available at ftp://ftp.ebi.ac.uk/pub/databases/embl/wgs/.
The genome monitoring table (MOT)
EBI's MOT (10) provides access to unfinished and finished genome data sorted by chromosome, enables navigation to individual EMBL-Bank entries and is updated daily at http://www.ebi.ac.uk/genomes/mot/.
Genome annotation and proteome analysis
The Ensembl Genome Browser provides the best possible automatic annotation, graphical views and web-searchable datasets for a number of eukaryotic genomes including human, mouse, drosophila, anopheles, zebrafish with others to follow. Automatic annotation, graphical views, web-searchable datasets including information on confirmed peptides, confirmed cDNAs, predicted peptides, repeat predictions along with integration of map information and SNPs are available from http://www.ensembl.org/.
Proteome Analysis information on a large number of organisms is available from SWISS-PROT at http://www.ebi.ac.uk/proteome/.
Sequence retrieval system (SRS)
The SRS server (11) at the EBI integrates and links a comprehensive collection of specialized databanks along with the main nucleotide and protein databases. Detailed instructions are available online at http://srs.ebi.ac.uk/.
Sequence searching
A comprehensive set of sequence similarity algorithms is available at URL: http://www.ebi.ac.uk/Tools/ or by Email. Users can search the database as a whole or by individual taxonomic division. The most Commonly used algorithms available are FASTA (12) and WU-BLAST (13). Comparisons between a nucleotide sequence and the protein databases can be made using fastx/y, while tfastx/y allows comparisons between a protein sequence and the translated DNA databank. The EBI's Smith and Watermann (14) service include MPsrch (reference—see help page), Edinburgh Biocomputing Systems (EBS) and Scanps (reference—see help page).
In total, more than 200 databases are available for searching at the EBI. The new fasta service for genomes and proteomes enables users to search on complete genomes and derived proteomes from public sequencing projects around the world.
Sequence analysis
Specialized sequence analysis programs include multiple sequence alignment and inference of phylogenies using CLUSTALW (15), Gene prediction using GeneMark (16), pattern searching and discovery using PRATT (17), Motif identification using ppsearch (see EBI's ppsearch help page) as well as applications which have been developed in-house for various other projects. EBI is in the process of adding more interactive sequence analysis resources based on the European Molecular Biology Open Software Suite (EMBOSS) (http://www.emboss.org/).
MAJOR NEW DEVELOPMENTS
New CON(struct) division
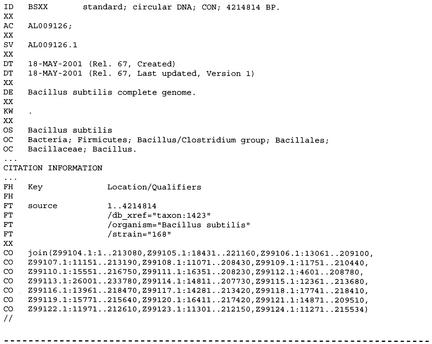
The new CON database division represents CON(structed) or CON(tig) sequences of chromosomes, genomes and other long sequences constructed from segment entries. Nucleotide sequence records in EMBL (as well as in DDBJ and GenBank) currently have a size restriction of 350,000 nucleotides (Fig. 1). Sequences >350 kb are split into smaller segment entries prior to inclusion in the database. Segment entries include sequence data, are assigned individual accession numbers and are distributed in the appropriate taxonomic divisions. In contrast, CON division entries do not contain sequence data per se, but rather the assembly information including all accession.versions and sequence locations relevant in building the contig sequence. CON sequence entries follow the daily data exchange mechanism between DDBJ/EMBL/GenBank.
Figure 1.
Relevant sections of the Bacillus subtilis CON entry providing construct information for the assembly of the B. subtilis bacterial genome (4.2 Mbases) from segment entries (<350 kb).
CON entries are available in the CON.dat file at ftp://ftp.ebi.ac.uk/pub/databases/embl/release/, from the EBI Genome Web server at URL http://www.ebi.ac.uk/genomes/ and also in the FTP Genomes directory at ftp://ftp.ebi.ac.uk/pub/databases/embl/genomes/, where CON entries are also complemented by EMBL flat-files including the complete DNA sequence and biological annotation. For an example see ftp://ftp.ebi.ac.uk/pub/databases/embl/genomes/Bacteria/bsubtilis/AL009126.embl. Furthermore, the complete CON sequence is made available in Fasta format, for an example see ftp://ftp.ebi.ac.uk/pub/databases/embl/genomes/Bacteria/bsubtilis/AL009126.fasta. Underlying segment entries are linked, searchable and retrievable via SRS and available for BLAST and FASTA homology searching.
Additional information is available from the EMBL User Manual at http://www.ebi.ac.uk/embl/Documentation/User_manual/co_line.html.
Whole genome shotgun (WGS) sequences
Methods using whole genome shotgun data are now used to gain a large amount of genome coverage for an organism. WGS data for O. sativa, A. gambiae, M. musculus, B. anthracis, Takifugu rubripes and many other organisms have been submitted to DDBJ/EMBL/GenBank. A complete list of WGS data currently available is maintained at http://www.ebi.ac.uk/genomes/wgs.html.
Example of WGS accession-number format: AAAA01000001.1
WGS accession-numbers consist of 〈4 letters〉〈2 digits〉〈6 digits〉.〈version〉
with,
〈4 letters〉=set_id
〈2 digits〉=set_version
〈6 digits〉=contig_id
〈version〉=sequence_version.
Oryza sativa L. ssp. indica WGS sequences have been assigned the project accession-number AAAA00000000. The first version of the project has the accession number AAAA01000000 and the first sequence in that set has acc#.version AAAA01000001.1 When a given WGS project is updated, ALL contigs from the first version are replaced by ALL contigs from the second version.
WGS data are available via EBI's SRS and from EBI's FTP server at ftp://ftp.ebi.ac.uk/pub/databases/embl/wgs.
Third Party Annotation (TPA) dataset
Following a decision taken at the 2002 Collaborative Meeting, DDBJ/EMBL/GenBank are in the process of creating a Third Party Annotation (TPA) dataset. Until now, the collaborative databases have built a comprehensive database of primary nucleotide sequences, resulting from direct sequencing of cDNAs, ESTs, genomic DNAs etc. Primary data are defined to be data for which the submitting group has done the sequencing and biological annotation, and as ‘owner’ of these data has privileges to submit updates/corrections etc. In contrast, non-primary sequences are defined as sequences which a) consist exclusively of DNA from one or several already existing entries ‘owned’ by other groups or b) consist of a mixture of new primary and already existing sequences.
Categories of data submissions that will be accepted for TPA include:
Re-annotation/analysis of sequence(s) from DDBJ/EMBL/GenBank.
Mixed primary/non-primary TPA sequence including regions of new and existing sequence (e.g. filling the gaps with HTG or EST or newly sequenced data).
TPA sequences based on trace sequences from the Ensembl/NCBI trace archive.
TPA sequences based on Whole Genome Shotgun data (WGS).
Not accepted are consensus sequences from multiple organisms.
Additional information required from TPA submitters
For TPA submissions, the database requires information on the composition of the TPA sequence to show, which spans in a TPA sequence originated from which contributing primary sequences. Webin-TPA prompts users to provide DDBJ/EMBL/GenBank accession-numbers/sequence versions and base-spans of the sequences used for the re-annotations/re-assemblies. If a TPA sequence includes a mixture of ‘old’ and ‘new’ sequence data, stretches of new primary sequence >300 bp first need to be submitted to the EMBL primary database (via WEBIN). In response, new database acc#s will be assigned and communicated, which can then be included in the list of contributing entries in the TPA record. For TPA sequences composed of raw sequences from the trace archive, the according trace archive identifiers (e.g. TI12234566) and spans need to be provided.
Information on contributing sequences will appear in the EMBL TPA record with new flat-file line-types AH/AS like in this example: 2 The Assembly Header (AH) line provides column headings for the assembly information. The AS (ASsembly) lines provide information on the composition of the TPA sequence by listing base span(s) of the TPA sequence together with identifiers and base spans of contributing sequences. Further details on new lines types are available at http://www.ebi.ac.uk/embl/Documentation/User_manual/usrman.html.
| AH | TPA-SPAN | PRIMARY_IDENTIFIER | PRIMARY_SPAN | COMP |
| AS | 1–426 | AC004528.1 | 18665–19090 | |
| AS | 427–526 | AC001234.2 | 1–100 | c |
Release policy
In order to assure that the sequence annotation is of high quality, we will be requiring, that the study has been published in a per-reviewed journal before we release the data to the public.
TPA data exchange
TPA sequences are exchanged amongst the DDBJ/EMBL/GenBank database collaboration on a daily basis, as a complement to the existing primary DDBJ/EMBL/GenBank database.
TPA data access
The TPA data collection is available via the EBI FTP server at ftp://ftp.ebi.ac.uk/pub/databases/embl/tpa and also via EBI's SRS at http://srs6.ebi.ac.uk.
Sequence length limits
Currently database records are limited in length to 350 kb. At the collaborative meeting in May 2002 DDBJ/EMBL/GenBank discussed the issue of relaxing the maximum sequence length limit. The plan is to remove the size restriction on database records in 2 years time. We will review this proposal in 12 months time.
EMBL Sequence Version Archive.
In order to provide access to previous versions of database records, the EMBL database has created Sequence Version Archive. Data in the EMBL nucleotide sequence database change over time for a number of reasons, e.g. due to updates/corrections or extensions based on new findings from more recent experiments. Each time data in an entry are modified, the entry is assigned a new entry version number.
But, an entry can change its appearance even while the data included remain unchanged, for example due to a general flat-file format change or when the taxonomic classification of the source organism changes, e.g. when an organism is assigned a new place in the hierarchy. Following these types of changes, the entry will retain its original version number.
Querying the EMBL Sequence Version Archive
The EMBL Sequence ‘Version’ Archive is available from the EBI web-servers at URL http://www.ebi.ac.uk/embl/sva. Entry(ies) can be viewed by either Accession number/Nucleotide Sequence identifier/Protein identifier.
Query results options will allow to:
Show the complete history for an entry, i.e. all recorded flat files matching the query criterion in chronological order or
Show a snapshot of the entry at a particular date. Query results will be presented in a table, listed EMBL entries can be ‘Viewed’ on Screen' or ‘Saved to File’.
Show differences between entry versions.
CITING THE EMBL DATABASE
The preferred form for citation of the EMBL Nucleotide Sequence Database is: Stoesser, G. et al. (2003) The EMBL Nucleotide Sequence Database. Nucleic Acids Res., 31, 1–6.
CONTACTING THE EMBL DATABASE
Computer network: data submissions datasubs@ebi.ac.uk; other inquiries, datalib@ebi.ac.uk; updates/publication notifications, update@ebi.ac.uk. Postal address: EMBL Nucleotide Sequence Submissions, European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SD, UK. Tel: data submissions, +44 1223494499; general, +44 1223494444. Fax: data submissions, +44 1223494472; general, +44 1223494468.
REFERENCES
- 1.Emmert D.B., Stoehr,P.J., Stoesser,G. and Cameron,G.N. (1994) The European Bioinformatics Institute (EBI) databases. Nucleic Acids Res., 22, 3445–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apweiler R., Attwood,T.K., Bairoch,A., Bateman,A., Birney,E., Biswas,M., Bucher,P., Cerutti,L., Corpet,F., Croning,M.D.R., Durbin,R., Falquet,L., Fleischmann,W., Gouzy,J., Hermjakob,H., Hulo,N., Jonassen,I., Kahn,D., Kanapin,A., Karavidopoulou,Y., Lopez,R., Marx,B., Mulder,N.J., Oinn,T.M., Pagni,M., Servant,F., Sigrist,C.J.A. and Zdobnov,E.M. (2001) interPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic. Acids Res., 29, 7–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boutselakis H., Dimitropoulos,D., Fillon,J., Golovin,A., Henrick,K., Hussain,A., Ionides,J., John,M., Keller,P.A., Krissinel,E., Mcneil,P., Naim,A., Newman,R., Oldfield,T., Pineda,J., Rachedi,A., Roser-Copeland,J., Sitnov,A., Sobhany,S., Suarez-Uruena,A., Swaminathan,J., Tagari,M., Tate,J., Tromm,S., Velankar,S. and Vranken,W. (2003) E-MSD: the European Bioinformatics Institute Macromolecular Structure Database. Nucleic Acids Res., 31, 458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brazma A., Sarkans,U., Robinson,A., Vilo,J., Vingron,M., Hoheisel,J. and Fellenberg,K. (2002) Microarray data representation, annotation and storage. Adv. Biochem. Eng. Biotechnol., 77, 113–139. [DOI] [PubMed] [Google Scholar]
- 6.Hubbard T., Barker,D., Birney,E., Cameron,G., Chen,Y., Clark,L., Cox,T., Cuff,J., Curwen,V., Down,T., Durbin,R., Eyras,E., Gilbert,J., Hammond,M., Huminiecki,L., Kasprzyk1,A., Lehvaslaihol,H., Lijnzaad,P., Melsopp,C., Mongin,E., Pettett,R., Pocock,M., Potter,S., Rust,A., Schmidt,E., Searle,S., Slater,G., Smith,J., Spooner,W., Stabenau,A., Stalker,J., Stupka,E., Ureta-Vidal,A., Vastrik,I. and Clamp,M. (2002) The Ensembl genome database project. Nucleic Acids Res., 30, 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tateno Y., Miyazaki,S., Ota,M., Sugawara,H. and Gojobori,T. (2000) DNA Data Bank of Japan (DDBJ) in collaboration with mass sequencing teams. Nucleic Acids Res., 28, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J., Rapp,B.A. and Wheeler,D.L. (2000) GenBank. Nucleic Acids Res., 28, 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lombard V., Camon,E.B., Parkinson,H.E., Hingamp,P., Stoesser,G., Redaschi,N. (2002) EMBL-Align: a new public nucleotide and amino acid multiple sequence alignment database. Bioinformatics, 18, 763–764. [DOI] [PubMed] [Google Scholar]
- 10.Beck S. and Sterk,P. (1998) Genome-Scale DNA sequencing where are we? Curr. Opin. Biotechnol., 9, 116–121. [DOI] [PubMed] [Google Scholar]
- 11.Zdobnov E.M., Lopez,R., Apweiler,R. and Etzold,T. (2002) The EBI SRS server—new features. Bioinformatics, 18, 1149–1150. [DOI] [PubMed] [Google Scholar]
- 12.Pearson W.R. (1994) Using the FASTA program to search protein and DNA sequence databases. Methods Mol. Biol., 24, 307–331. [DOI] [PubMed] [Google Scholar]
- 13.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 14.Smith R.F. and Waterman,M.S. (1981) Comparison of biosequences. Adv. Appl. Math., 2, 482–489. [Google Scholar]
- 15.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borodovsky M. and McIninch,J. (1993) GeneMark: Parallel Gene Recognition for both DNA Strands. Comput. Chem., 17, 123–133. [Google Scholar]
- 17.Jonassen I., Collins,J.F. and Higgins,D.G. (1995) Finding flexible patterns in unaligned protein sequences. Protein Sci., 4, 1587–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]