Abstract
The Adenylate Uridylate (AU)-Rich Element Database, ARED-mRNA version 2.0, contains information not present in the previous ARED. This includes additional data entries, new information and links to Unigene, LocusLink, RefSeq records and mouse homologue data. An ARE consensus sequence specific to the 3′UTR is the basis of ARED that demonstrated two important findings: (i) AREs are present in a large, previously unrecognized set of human mRNAs; and (ii) ARE-mRNAs encode proteins of diverse functions which are largely involved in early and transient biological responses. In this update, we have modified the strategy for identifying ARE-mRNA in order to systematically deal with inconsistencies of molecule type and mRNA region in GenBank records. Potential uses for the ARED in functional genomics are also given. The database is accessible via the web, http://rc.kfshrc.edu.sa/ared, with a new querying system that allows searching ARE-mRNAs by any public database identifier or name. The ARED website also contains relevant links to uses for the ARED.
INTRODUCTION
Messenger RNA turnover is an important process involved in the transient response to microbes and stress agents. Functionally defined and derived adenylate–uridylate rich element (ARE) consensus sequences have been shown to exist in the 3′UTR of selected mRNAs belonging to interferons, cytokines and proto-oncogenes (1). A 13-bp ARE motif was computationally derived from a list of functionally labile ARE-mRNAs and was the basis of the ARE-mRNA database (ARED) which contains GenBank entries where the 3′UTR matches the motif (2). The ARED demonstrated that ARE-mRNAs represent as much as 5–8% of human genes and encode functionally diverse proteins that are important in many transient biological processes including cell growth and differentiation, signal transduction, transcriptional and translational control, hematopoiesis, apoptosis, nutrient transport, and metabolism (2). Here, we further expand ARED using a more comprehensive text extraction approach while systematically dealing with GenBank inconsistency in molecule type and region of mRNA, and present examples of the potential applications of AREs in functional genomics.
METHODS
The mRNA molecular type, mRNA region and dealing with GenBank inconsistencies: examples
The aim of the database is to search for ARE-mRNAs (cDNAs) using 3′ UTR-specific ARE consensus sequences. Our search strategy uses a comprehensive approach for extracting mRNA (cDNA) with full-length coding regions while dealing with inconsistencies in GenBank in regard to nucleic acid molecule type. For example, we have avoided solely searching for records with molecule type, mRNA or RNA, in the LOCUS field or searching solely for mRNA or cDNA in the DEFINITION line. During our searches, we have observed that there are genuine mRNA records but defined incorrectly as DNA instead of mRNA or RNA in the LOCUS field. For example; DEFINITION: Homo sapiens mRNA for thromboplastin; LOCUS: DNA; ACC=A19048. In many records, gene in the DEFINITION line refers to a gene, e.g. with introns and exons, such as DEFINITION: H. sapiens CC chemokine LCC-1 precursor, gene, Locus: DNA, ACC=AF039954; the molecule type in the DEFINITION line described as gene may actually refer to an mRNA molecule, for example, DEFINITION: Human gene for fibroblast (beta-1) interferon, Locus: DNA in which the true molecule type is mRNA (FEATURE table). We did not rely on finding complete CDS through the DEFINITION line [Example: DEFINITION: Human interleukin 1-beta (IL1B) mRNA, complete cds, ACC=M15330], since there are records that have complete CDS in their Feature but not described in Definition line; for example, DEFINITION: Human mRNA for tumor necrosis factor. ACC=X01394. Also, not all the 3′UTR regions are defined in FEATURE key (3); thus, we computationally extracted 3′UTR (>CDS in FEATURE).
ARED 2.0 buildup
To avoid the above inconsistencies, we first comprehensively extract all possible records as shown in Figure 1 and followed by exclusion of records that distinguish molecule type or mRNA region such as words like introns, HTG, PAC (Fig. 1). Subsequently, computational extraction of 3′UTR and removal of redundancy was performed. Redundancy was further refined by consolidation with UniGene and RefSeq databases. The 3′UTRs were searched for the 13-bp pattern WWWUAUUUAUWW with mismatch=−1 which was computationally derived as previously described (2). The pattern was further statistically validated against larger sets of mRNA data (10 872 mRNA with 3′UTR; GenBank 119) showing occurrence of the motif in 6.8% of human mRNA. When compared to other regions of the mRNA, 5′UTR and CDS; the specificity to 3′UTR was 99.4% (P<0.0001) and 98.9% (P<0.0001), respectively. The statistical methodology was previously described (2).
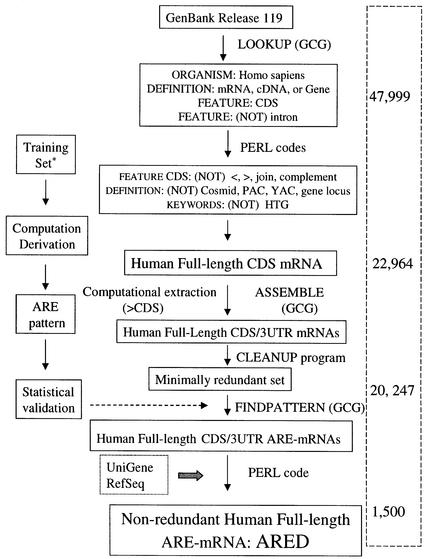
Figure 1.
ARED 2.0 buildup strategy. A schematic chart showing the stages of theARED 2.0 buildup. Programs and *computational derivation of ARE motif used in this study has been previously described (2). Programs are either PERL scripts or part of the GCG (Wisconsin Bioinformatics Package). Numbers on the right column indicate number of sequences.
THE DATABASE AND BIOLOGICAL APPLICATIONS
The diversity of biological processes represented in the ARED allows multiple applications in several fields of interest. It is possible to search the database for any gene of interest to see whether it belongs to the ARED or any of its cluster groups (4–6). The latter is based on the length of the ARE stretch and may help distinguish between functionally typical (which are probably have longer stretch) and atypical ARE-mRNA.
A cDNA microarray was used for the measurement of mRNA turnover in induced B-cell lymphoma and peripheral blood mononuclear cells (7). Using ARED, the authors concluded that our computationally extracted ARE motif was preferentially found in the most unstable mRNA (<2 h) and was observed with decreasing frequency in more stable mRNAs (>8 h). This is opposite to that found in non-ARE genes in which stable mRNA constitutes the majority (60%) of mRNAs observed in this study (7).
Another example of the utility of the ARED is our integrated computational and laboratory approach for selective amplification of ARE-mRNAs (8) in which statistical analysis of the initiation regions in the 5′UTR of ARE-mRNAs was performed and accordingly, several 5′ primers and a single universal 3′ primer that targeted the initiation consensuses and ARE regions, respectively, were designed. This resulted in selective amplification of ARE-mRNA, many of which are present in ARED. Finally, we have used the ARED to create a custom cDNA array containing 950 ARE-mRNAs and investigated mRNA induction and decay in a reproducible cellular model system. This has allowed us to identify a positive feedback loop regulating transcript stabilization and the association of long stretches of AREs with labile mRNAs (9).
ARE-mediated changes in mRNA stability are important in processes that require transient responses such as cellular growth, immune response, cardiovascular toning and external stress-mediated pathways. Stabilization of the ARE-mRNAs can cause prolonged responses that subsequently may lead to undesirable effects e.g. diseased states. Indeed, several diseases including certain cancer states and chronic inflammatory conditions (2) are known to be caused by stabilized ARE-mRNAs. Thus, the ARED may contribute further to our understanding of the relationships between AREs and certain diseases.
THE ARED WEBSITE AND FUTURE PERSPECTIVES
The ARED website (http://rc.kfshrc.edu.sa/ared) offers a query search engine that allows searches for ARE-genes using multiple identifier numbers or descriptions such as UniGene IDs, UniGene definition, RefSeq IDs, accession numbers, alternative names, official Gene symbols and mouse homologs (MGD) (10). The ARE-mRNAs were also clustered based on the lengths of the AREs with Cluster Group I contain 5 continuous AREs, Group/II containing 4 continuous AREs, Group III contains three continuous AREs, Group IV contain 2 continuous AREs and finally Group V contains one ARE in a 13-bp ARE context; the clusters can also be queried. The database is available as a single GenBank flat file (i.e. nucleotide sequence with annotations) that can be requested from the authors. We are currently focusing on the functional genomics of AREs to establish global relationships between the computational and functional elements.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Dr Sayeda Abu-Amero for reviewing the manuscript. Shazia Subhani, Bushra Siddiqui and Noel De Leon, Department of Biostatistics Epidemiology, and Scientific Computing (KFSH&RC) for database search engine and web development.
REFERENCES
- 1.Chen C.Y. and Shyu,A.B. (1995) AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci., 20, 465–470. [DOI] [PubMed] [Google Scholar]
- 2.Bakheet T., Frevel,M., Williams,B.R.G., Greer,W. and Khabar,K.S.A. (2001) ARED: Human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional reportiore of encoded proteins. Nucleic Acids Res., 29, 246–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalphin M.E., Stockwell,P.A., Tate,W.P. and Brown,C.M. (1999) TransTerm, the translational signal database, extended to include full coding sequences and untranslated regions. Nucleic Acids Res., 27, 293–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marion E., Kaisaki,P.J., Pouillon,V., Gueydan,C., Levy,J.C., Bodson,A., Krzentowski,G., Daubresse,J.C., Mockel,J., Behrends,J., Servais,G., Szpirer,C., Kruys,V., Gauguier,D. and Schurmans,S. (2002) The gene INPPL1, encoding the lipid phosphatase SHIP2, is a candidate for type 2 diabetes in rat and man. Diabetes, 51, 2012–2017. [DOI] [PubMed] [Google Scholar]
- 5.Rundlof A.K., Carlsten,M. and Arner,E.S. (2001) The core promoter of human thioredoxin reductase 1: cloning, transcriptional activity, and Oct-1, Sp1 and Sp3 binding reveal a housekeeping-type promoter for the AU-rich element-regulated gene. J. Biol. Chem., 276, 30542–30551. [DOI] [PubMed] [Google Scholar]
- 6.Gouble A., Grazide,S., Meggetto,F., Mercier,P., Delsol,G. and Morello,D. (2002) A new player in oncogenesis: AUF1/hnRNPD overexpression leads to tumorigenesis in transgenic mice. Cancer Res., 62, 1489–1495. [PubMed] [Google Scholar]
- 7.Lam L.T. (2001) Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol., 2, research0041.1–0041.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khabar K.S., Dhalla,M., Bakheet,T., Sy,C. and al-Haj,L. (2002) An integrated computational and laboratory approach for selective amplification of mRNAs containing the adenylate uridylate-rich element consensus sequence. Genome Res., 12, 985–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frevel M.A., Bakheet,T., Silva,A.M., Hissong,J., Khabar,K.S. and Williams,B.R. (2002) P38 MAP kinase dependent and independent signaling of mRNA stability of AU-rich element containing transcripts. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blake J.A., Richardson,J.E., Bult,C.J., Kadin,J.A. and Eppig,J.T. (2002) The Mouse Genome Database (MGD): the model organism database for the laboratory mouse. Nucleic Acids Res., 30, 113–115. [DOI] [PMC free article] [PubMed] [Google Scholar]