Abstract
We describe here the establishment of an online database containing a large number of sequences and related data on viroids, viroid-like RNAs and human hepatitis delta virus (vHDV) in a customizable and user-friendly format. This database is available on the World Wide Web at http://penelope.med.usherb.ca/subviral.
INTRODUCTION
Viroids, plant satellite viroid-like RNAs, and human hepatitis delta virus (vHDV) form the ‘brotherhood’ of the smallest known auto-replicable RNAs. Since 1996, we have maintained an online database of auto-replicable RNAs, in order to facilitate research on these species by presenting a large number of sequences and related data in a comprehensive and user-friendly format [e.g. position of their self-catalytic domains, the open reading frame for vHDV, prediction of the most stable secondary structures, etc. (1–5)]. In this report, we describe a completely re-engineered subviral RNA database which includes more sequences and features which facilitate and customize the data retrieval process.
DESCRIPTION OF THE SUBVIRAL RNA DATABASE
The SubViral RNA database comprises all sequences, to our knowledge, that have been either published or were available from sequence library file servers. Upon entering into the database, one encounters a menu which lists the different accessible sections (i.e. Viroids, Satellite RNAs, vHDV and Related RNAs). These sections are always available so as to allow easy access to any section. The choice of a section leads to the second level, which is a summary table for each sequence subdivision. Briefly, each species of RNA is listed by its complete name, its abbreviation, the number of sequence variants and its size distribution. The third level is entered by choosing a particular species and leads to a complete listing of the sequence variants named by their assigned nomenclature. In order to simplify the nomenclature of the included sequences, we used an identification scheme based on the usual abbreviation of an RNA species followed by a number (1). Their accession number, self-cleaving motif (if present), and their sequence length and composition are also listed. The fourth level contains additional data for each chosen entry, including their accession numbers, bank loci (when available), number of nucleotides (total and by type), complete publication information and the sequence in 10 nt blocks. In addition, secondary structure predictions of the most likely ancestral variants are appended to the database. The analysis of the viroid and viroid-like RNA sections (e.g. classification, secondary structure prediction, phylogenetic identification of the likely ancestral variant, etc.) has been the object of a previous report (1).
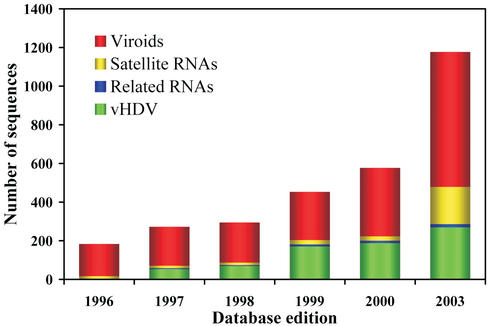
Today more than 1200 sequences are indexed in the database, more than six times the number present in the original edition. Since the original release, the number of sequences has increased swiftly and considerably (Fig. 1). Our catalogue comprises 66 species containing 698 sequences from viroids, 193 from plant satellite viroid-like RNAs, 16 from related species of RNA and 270 complete or partial sequences of vHDV.
Figure 1.
Evolution of the database since its creation. The number of sequences indexed for each section of the database was taken from each database edition published.
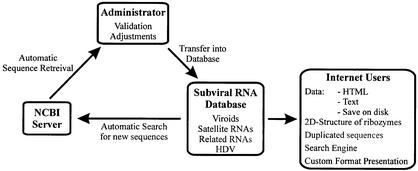
The 2002 edition of the SubViral RNA database has been completely re-engineered. From a primarily hypertext content, most of the data have been transferred to a MySQL database server. Each entry in the database is now accessible via a CGI script running on the server. This architecture maximizes the speed and permits a superior customizability of the database. Moreover, sequences are automatically added on a daily basis from the sequence library file servers (NCBI sequence libraries). A schematic representation of the data retrieval/presentation process is shown in Figure 2. In order to ensure homogeneity, each new sequence entry is manually validated and adjusted (e.g. all sequences from a species are adjusted so as to have the same sequence origin and polarity).
Figure 2.
Organigram representation of the database operation.
On top of the above improvements, several new features have been added. It is now possible to select entries, individually or in groups, and to display them in a customizable format (e.g. complete information, FASTA format, selected fields, etc.). For sequences that possess self-catalytic hammerhead and hairpin structures, their nucleotide sequences and secondary structures are presented. A search engine, which permits the user to query through the database, has also been added. Duplicated sequences are now presented for each entry, providing a better overview of the sequences diversity. Secondary structure predictions of the delta ribozyme will be added soon. Finally, more information on these pathogenic RNAs, and on the cleavage efficiencies of self-cleaving motifs, will be added in the near future.
The SubViral RNA database is available on the World Wide Web at the URL http://penelope.med.usherb.ca/subviral. The database is updated automatically as soon as sequences become available, and contains more entries than are present in the GenBank and EMBL nucleotide sequence libraries. Users of the viroid and viroid-like RNA database should cite this publication, and are encouraged to provide corrections, or other information, for inclusion in the database via electronic mail (map@penelope.med.usherb.ca).
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Dr Robert A. Owens for maintaining the databases for the last two years. This work was supported by a grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada to J.-P.P. The RNA group is supported by grants from both the Canadian Institutes of Health Research (CIHR) and Fonds FCAR (Québec). M.P. is the recipient of a post-doctoral fellowship from the CIHR. J.P. is the recipient of a predoctoral fellowship from the Fonds de la Recherche en Santé du Québec (FRSQ). J.-P.P. is an Investigator of the CIHR.
REFERENCES
- 1.Bussière F., Lafontaine,D. and Perreault,J.P. (1996) Compilation and analysis of viroid and viroid-like RNA sequences. Nucleic Acids Res., 24, 1793–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lafontaine D., Mercure,S. and Perreault,J.P. (1997) Update of the viroid and viroid-like sequence database: addition of a hepatitis delta virus RNA section. Nucleic Acids Res., 25, 123–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lafontaine D.A., Mercure,S., Poisson,V. and Perreault,J.P. (1998) The viroid and viroid-like RNA database. Nucleic Acids Res., 26, 190–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lafontaine D.A., Deschênes,P., Bussière,F., Poisson,V. and Perreault,J.P. (1999) The viroid and viroid-like RNA database. Nucleic Acids Res., 27, 186–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pelchat M., Deschênes,P. and Perreault,J.P. (2000) The database of the smallest known auto-replicable RNA species: viroids and viroid-like RNAs. Nucleic Acids Res., 28, 179–180. [DOI] [PMC free article] [PubMed] [Google Scholar]