Abstract
The Zebrafish Information Network (ZFIN) is a web based community resource that serves as a centralized location for the curation and integration of zebrafish genetic, genomic and developmental data. ZFIN is publicly accessible at http://zfin.org. ZFIN provides an integrated representation of mutants, genes, genetic markers, mapping panels, publications and community contact data. Recent enhancements to ZFIN include: (i) an anatomical dictionary that provides a controlled vocabulary of anatomical terms, grouped by developmental stages, that may be used to annotate and query gene expression data; (ii) gene expression data; (iii) expanded support for genome sequence; (iv) gene annotation using the standardized vocabulary of Gene Ontology (GO) terms that can be used to elucidate relationships between gene products in zebrafish and other organisms; and (v) collaborations with other databases (NCBI, Sanger Institute and SWISS-PROT) to provide standardization and interconnections based on shared curation.
INTRODUCTION
The zebrafish has emerged as a model organism important for the identification and characterization of genes and pathways involved in development, organ function, behavior and disease. One of the many challenges produced by the rapid expansion of zebrafish research is to provide a centralized resource for the curation and integration of zebrafish data and their integration with data from other model organisms and humans. ZFIN fills this role by providing searchable interfaces to access mutants, genes, genetic markers, mapping panels, links to other genomic resources, publications and community contact data (1). Data integration fosters an understanding of gene function by linking genotypes, phenotypes and gene expression to gene sequences and gene models. ZFIN is continually expanded through ongoing literature curation and bulk data loads.
ANATOMICAL DICTIONARY
ZFIN serves as the focal point for the development and the dissemination of the zebrafish anatomical dictionary. The dictionary organizes anatomical terms, for a series of developmental stages and the adult, in a hierarchical manner. The dictionary provides a standard vocabulary for annotating gene expression and phenotype thus providing a link between these two types of data commonly used to study gene function. The use of standard anatomical nomenclature enables comparisons of data among organisms. Anatomical dictionary terms may be used to query gene expression data found in ZFIN (http://zfin.org/cgi-bin/webdriver?MIval=aa-xpatselect.apg).
GENE EXPRESSION
ZFIN provides an integrated analysis of genotype, phenotype and expression data through access to data from mRNA in situ hybridizations. These data are annotated using keywords and developmental stages from the zebrafish anatomical dictionary. Researchers may perform complex searches of these data to find the expression patterns of genes or gene families. Based on gene name, developmental stages, anatomical terms and genetic map positions ZFIN displays images as well as text summaries describing the expression data (Fig. 1).
Figure 1.
RNA in situ hybridization assay for the neurogenin 1 (neurog1) gene from 50%-epiboly through Bud developmental stages of the gastrula. Patterns are grouped by stage. Anatomical dictionary terms identify regions of gene expression. A text description describes observed patterns. Links are provided to relevant ZFIN gene record and stage descriptions.
SEQUENCE
The zebrafish whole-genome sequencing initiative began in February 2001 at the Sanger Institute. ZFIN and the Sanger Institute collaborate to identify locations of known genes and markers and to identify novel genes. The long term plan is for ZFIN to serve as the repository for this information, integrating annotated sequence with ZFIN genotype, phenotype and gene expression data. To this end, ZFIN has begun to create an infrastructure that will take advantage of the genome sequence information. Curator annotated EMBL records of molecular segments sequenced and submitted by the Sanger Institute are incorporated into ZFIN and linked to other ZFIN objects. Researchers may query ZFIN for molecular segment data by name, sequence type, markers contained within the segment and genetic map locations. ZFIN displays relationships between genes and clones, sequence similarities for novel genes, information about the clones and links to the corresponding sequences.
GENE ONTOLOGY
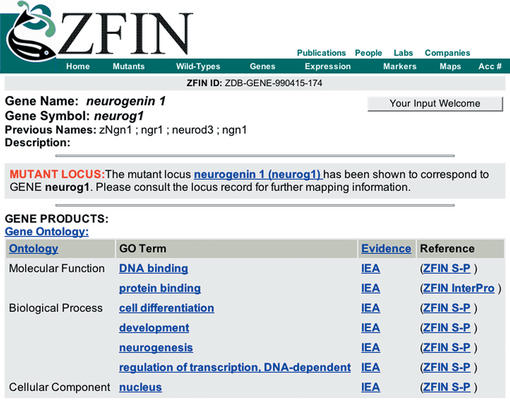
One of ZFIN's major goals is to add information about gene products in a manner that will aid in the elucidation of relationships among genes in zebrafish and between zebrafish and other organisms. To this end, ZFIN has begun automated annotation of gene records with Gene Ontology (GO; http://geneontology.org) terms, a dynamic controlled vocabulary developed by the gene ontology consortium that can be applied to all organisms even as knowledge of gene and protein roles in cells is accumulating and changing (2). Thus far, ontologies describing molecular function, biological process and cellular component have been defined. GO annotations (Fig. 2) are assigned to ZFIN gene records based on an automated association of SWISS-PROT keywords and InterPro records with GO terms using translation tables made available by GO. A collaboration with the SWISS-PROT database group enables ZFIN to maintain robust links to corresponding SWISS-PROT records. GO terms are linked to the QuickGO page describing the term's lineage in the ontology. GO defined evidence codes are displayed to allow researchers to assign a level of confidence to the annotation of a gene by a particular GO term. A link to the reference supporting the annotation is also provided.
Figure 2.
GO annotation display for the neurog1 zebrafish gene. GO terms are grouped by ontologies. Evidence codes and references provide researchers with information to establish levels of confidence for the association. GO term links provide information describing the lineage of the GO term.
COLLABORATIONS
ZFIN participates in collaborations with NCBI, the Sanger Institute and SWISS-PROT by exchanging manually curated data. Through these collaborations, nucleic acid sequence, protein, and InterPro protein domain links are added to ZFIN gene records. Such links increase the robustness of associations of genes with sequences.
USER INPUT
ZFIN recognizes the importance of community input to its success. ZFIN encourages new data submissions and comments on its annotation efforts through a ‘Your input welcome’ button displayed on ZFIN pages. General comments, questions and suggestions may be sent to zfinadmn@zfin.org.
IMPLEMENTATION
ZFIN is implemented with the IBM/Informix relational database management system (version 9.21). A web interface of HTML-based forms combined with JavaScript, Java, Perl and CGI scripts provides access to the database. The current ZFIN data model may be viewed at http://zfin.org/zf_info/dbase/PAPERS/CurrentDataModel/db.html.
Acknowledgments
ACKNOWLEDGEMENT
Funds for the development of the Zebrafish Information Network are provided by the NIH (RR/HD 12546).
REFERENCES
- 1.Sprague J., Doerry,E., Douglas,S., Westerfield,M. and the ZFIN Group (2001) The Zebrafish Information Network (ZFIN): a resource for genetic, genomic and developmental research. Nucleic Acids Res., 29, 87–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Gene Ontology Consortium (2001) Creating the Gene Ontology Resource: design and implementation. Genome Res., 11, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]