Abstract
The SeedGenes database (http://www.seedgenes.org) presents molecular and phenotypic information on essential, non-redundant genes of Arabidopsis that give a seed phenotype when disrupted by mutation. Experimental details are synthesized for efficient use by the community and organized into two major sections in the database, one dealing with genes and the other with mutant alleles. The database can be queried for detailed information on a single gene to create a SeedGenes Profile. Queries can also generate lists of genes or mutants that fit specified criteria. The long-term goal is to establish a complete collection of Arabidopsis genes that give a knockout phenotype. This information is needed to focus attention on genes with important cellular functions in a model plant and to assess from a genetic perspective the extent of functional redundancy in the Arabidopsis genome.
DATABASE DESCRIPTION
Seed development has become a popular subject for genetic analysis because it plays a critical role in the life cycle of flowering plants (1). Many genes must be expressed as the zygote divides in a regulated manner, completes morphogenesis and differentiates into a mature embryo. We have chosen to focus on essential, non-redundant genes that give a seed phenotype when disrupted by mutation. These genes represent an important component of the minimal gene set required to make a functional plant. We are studying genes that give either an embryo-defective or a seed pigment phenotype when disrupted by a loss-of-function mutation. Arabidopsis appears to contain about 750 such genes required for normal seed development (2,3). Several recent studies have dealt with the identification of essential genes in the unicellular Saccharomyces cerevisiae (4) and the multicellular Caenorhabditis elegans (5,6). Our goal is to establish a foundation for related studies in a model plant.
The SeedGenes project is a collaboration between the Meinke laboratory at Oklahoma State University, Syngenta in North Carolina and the Virginia Bioinformatics Institute. This project is an extension of longstanding efforts to screenT-DNA insertion lines for seed-defective mutants, resolve tagging status through genetic analysis and recover flanking sequences of tagged mutants using TAIL-PCR (3). The mutant population we are using is distinct from the one designed for reverse genetics at Syngenta and made available to the community through the Torrey Mesa Research Institute. First time users of the project database must register and provide basic information for project tracking purposes. For all subsequent logins, only a registered email address is required.
The primary objective of the SeedGenes project is to establish a foundation for identifying every Arabidopsis gene with an essential function during seed development. We are coordinating the collection, analysis and presentation of information on these genes based on cloning of mutant alleles. Information on essential genes and their corresponding mutants is obtained from both public and private sources and is then synthesized for efficient use by the community. We anticipate that individuals with an interest in seed development will utilize this database to keep track of the full spectrum of genes known to be required at this critical stage. Plant biologists can visit the site to determine if their favorite gene of interest performs an essential function in growth and development. In cases where only weak mutant alleles have been studied in the past, the database could be consulted to determine if a null allele results in embryo abortion. Tutorials planned for future releases should make it easier for members of the Arabidopsis community to screen their own knockout lines for a mutant seed phenotype and determine if a failure to obtain homozygotes is due to embryo lethality. The database should also help to focus attention on plant proteins with unknown functions that merit further analysis because they are known to be essential. One practical application of the database will be to identify a number of related genes with important roles in seed development in crop plants.
Project deliverables over the next 3 years include public access to information and seed stocks for 500 mutants defective in 300 different EMB genes; similar information for another 100 pigment mutants defective in 75 genes; expression data for genes active in young seeds; and a database that should serve as a model for presenting synthesized information on large collections of mutants. The first version of the database released in March 2002 contained 101 genes and 143 mutants, including those described in existing research publications. Target numbers for future releases are outlined in Table 1. The number of mutants exceeds the number of genes because in many cases duplicate alleles are involved. Mutants and genes have initially come from Syngenta and the Arabidopsis community at large, with additional partners expected in future releases. All seed stocks are being made available through the Arabidopsis Biological Resource Center (ABRC). Syngenta has agreed to provide gene identities and seed stocks derived from their internal research programs without involvement of material transfer agreements, reach-through rights, or requirement for written notification of pertinent publications or future inventions. We believe this represents a positive model for public release of extensive private sector resources.
Table 1. Timetable for addition of essential genes to the project database.
| Month | Year | Project months | Syngenta EMB | Other EMB | Seed pigment | Total genes |
|---|---|---|---|---|---|---|
| September | 2001 | 0 | — | — | — | — |
| March | 2002 | 6 | 50 | 25 | 25 | 100 |
| September | 2002 | 12 | 100 | 40 | 30 | 170 |
| March | 2003 | 18 | 150 | 50 | 40 | 240 |
| September | 2003 | 24 | 165 | 65 | 50 | 280 |
| March | 2004 | 30 | 180 | 80 | 60 | 320 |
| September | 2004 | 36 | 190 | 90 | 65 | 345 |
| March | 2005 | 42 | 195 | 95 | 70 | 360 |
| September | 2005 | 48 | 200 | 100 | 75 | 375 |
The SeedGenes ‘Query’ page can be used either to get detailed information on a given gene and its corresponding mutant alleles or to obtain lists of genes or mutants that match selected criteria. Examples of gene criteria include: gene class (embryo defective versus seed pigment), chromosome location and availability of a second mutant allele and full-length cDNA sequence. Genes can also be queried using a protein function keyword. Selection criteria for mutants include: original seed source, terminal phenotype class, mutagen type, tagging status, inheritance pattern and seed color.
THE SEEDGENES PROFILE
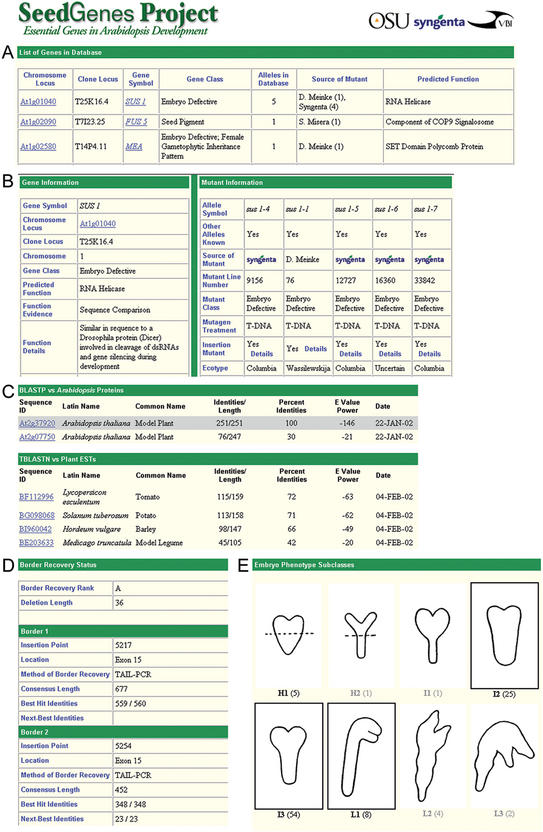
The cornerstone of the database is the Gene Profile feature. We worked to create a succinct page that contains the most important information about a gene and its mutant alleles. Each term included in the Profile is defined in a linked glossary. Additional details can be obtained through The Arabidopsis Information Resource (7). Highlights of selected components of the SeedGenes database are shown in Figure 1. Gene details in the Profile include: gene symbol, chromosome locus (e.g. At1g01040), clone locus (e.g. T25K16.4), gene class, predicted protein function, evidence in support of predicted function, certainty of gene identity, other gene identifiers, the predicted length and sequence of the gene and its products and top BLAST hits to proteins from Arabidopsis, translated plant ESTs and non-plant model organisms. In addition, Pfam motifs and predicted cellular localization based on TargetP are presented. An example of the BLAST summary is presented in Figure 1C.
Figure 1.
Representative components of the SeedGenes Project web page. (A) Portion of a gene list generated in response to a query; (B) portion of a SeedGenes Profile with gene information (left) and mutant information (right); (C) portion of a BLAST summary table; (D) border details for a T-DNA insertion mutant; and (E) diagram summarizing terminal phenotypes for an embryo-defective mutant.
The ‘Mutant’ section of the Profile includes details on seed source, mutagenic treatment, terminal phenotype, existence of duplicate mutant alleles, molecular location of the lesion within the gene, seed and embryo color, genetic segregation data and average seed and embryo lengths. The important contribution here is that seed mutants isolated in many different laboratories are characterized in the same manner with results summarized according to a standardized format. Links are available to additional pages that provide details on border recovery status for insertion mutants and terminal phenotype classes. A standardized classification system was developed to allow arrested embryos of many different shapes and sizes to be placed into terminal phenotype categories based on external morphology. Examples of several such classes are shown in Figure 1E. Detailed information on insertion alleles includes the vector name, selection agent, genetic evidence in support of tagging, length of deletion associated with the insertion and level of confidence that the correct gene has been identified. The length and insertion point for each sequenced border are presented along with the identity of the next most similar gene in the Arabidopsis genome in order to demonstrate that the correct gene has been identified.
DATABASE DESIGN
The SeedGenes architecture consists of a web interface produced by Java servlets running in a Tomcat server. The servlets handle queries and retrieve data from the database (Oracle 8i) using JDBC. The relational schema, developed using ERWin (Computer Associates, Inc.), attempts to balance normalization with query simplicity. The primary data flow into the database is from Excel spreadsheets containing genetic, molecular and phenotypic details. For the Syngenta collection of insertion mutants, most of the genetic and phenotypic data were generated in the Meinke laboratory whereas gene identities and insertion points were derived from research at Syngenta. For each database release, new data spreadsheets are transferred to the Dickerman group at VBI, where they are parsed into about 30 relational tables using Oracle PL/SQL scripting language. Tables with gene information such as predicted function, BLAST and TargetP results, alias symbols and identity evidence are linked through the unique AGI gene identifier (8), whereas tables with mutant information such as mutagenic treatment, segregation ratios, seed phenotypes and molecular insertion points are linked through a unique allele symbol. Both sets of tables are connected through a reference mutant allele that is associated with each gene included in the database.
FUTURE ENHANCEMENTS
Plans for future database releases include the incorporation of additional details on special embryo and endosperm defects observed in mutant seeds, along with selected images of cleared seeds viewed with Nomarski optics to provide a more complete picture of the mutant phenotype. Assignments of genes to functional classes will utilize the gene ontology system (9) when it becomes available for the entire Arabidopsis genome. Conclusions from gene expression experiments performed throughout the community and in association with the SeedGenes project will be added to the project database to provide additional details on the nature and diversity of genes transcribed during early stages of seed development. The project web site will be expanded to include a tutorial on seed development to help users screen their favorite gene knockouts for a seed phenotype. Together these modifications should facilitate the compilation of an extensive and informative list of genes with essential functions during reproductive development in Arabidopsis.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Colleen Sweeney, Rebecca Rogers, Steven Hutchens and Shkelzen Shabani for help with data processing at Oklahoma State University and Paul Toffenetti at VBI for assistance with database administration. Many additional contributors are listed on the SeedGenes web site. Development of the SeedGenes database was supported by the NSF 2010 Program and the S.R. Noble Foundation.
REFERENCES
- 1.Lohe A.R. and Chaudhury,A. (2002) Genetic and epigenetic processes in seed development. Curr. Opin. Plant Biol., 5, 19–25. [DOI] [PubMed] [Google Scholar]
- 2.Franzmann L.H., Yoon,E.S. and Meinke,D.W. (1995) Saturating the genetic map of Arabidopsis thaliana with embryonic mutations. Plant J., 7, 341–350. [Google Scholar]
- 3.McElver J., Tzafrir,I., Aux,G., Rogers,R., Ashby,C., Smith,K., Thomas,C., Schetter,A., Zhou,O., Cushman,M.A. et al. (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics, 159, 1751–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giaever G., Chu,A.M., Ni,L., Connelly,C., Riles,L., Veronneau,S., Dow,S., Lucau-Danila,A., Anderson,K., Andre,B. et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature, 418, 387–391. [DOI] [PubMed] [Google Scholar]
- 5.Fraser A.G., Kamath,R.S., Zipperlen,P., Martinez-Campos,M., Sohrmann,M. and Ahringer,J. (2000) Functional genomic analysis of C.elegans chromosome I by systematic RNA interference. Nature, 408, 325–330. [DOI] [PubMed] [Google Scholar]
- 6.Gonczy P., Echeverri,C., Oegema,K., Coulson,A., Jones,S.J., Copley,R.R., Duperon,J., Oegema,J., Brehm,M., Cassin,E. et al. (2000) Functional genomic analysis of cell division in C.elegans using RNAi of genes on chromosome III. Nature, 408, 331–336. [DOI] [PubMed] [Google Scholar]
- 7.Huala E., Dickerman,A.W., Garcia-Hernandez,M., Weems,D., Reiser,L., LaFond,F., Hanley,D., Kiphart,D., Zhuang,M., Huang,W. et al. (2001) The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res., 29, 102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 9.The Gene Ontology Consortium (2001) Creating the gene ontology resource: design and implementation. Genome Res., 11, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]