Abstract
The PRINTS database houses a collection of protein fingerprints. These may be used to assign uncharacterised sequences to known families and hence to infer tentative functions. The September 2002 release (version 36.0) includes 1800 fingerprints, encoding ∼11 000 motifs, covering a range of globular and membrane proteins, modular polypeptides and so on. In addition to its continued steady growth, we report here the development of an automatic supplement, prePRINTS, designed to increase the coverage of the resource and reduce some of the manual burdens inherent in its maintenance. The databases are accessible for interrogation and searching at http://www.bioinf.man.ac.uk/dbbrowser/PRINTS/.
INTRODUCTION
Fingerprints are groups of conserved sequence motifs that together provide diagnostic signatures for protein families. They derive much of their potency from the context afforded by multiple-motif matching, making them more flexible and powerful than single-motif approaches. Unlike some other pattern-matching methods, fingerprinting is well-suited to the creation of ‘hierarchical’ discriminators—e.g. this approach has been used to resolve G protein-coupled receptor (GPCR) super-families into their constituent families and receptor sub-types (1), and to sub-classify a variety of channel proteins, transporters and enzymes.
To date, 1800 fingerprints have been developed, manually annotated and deposited in the PRINTS database (2). Overall, the database is still rather small, largely because detailed annotation of entries is extremely time-consuming. However, the extent of manually-crafted annotations sets the database apart from the growing number of automatically-derived ‘family’ resources, for which there is no biological documentation and no result validation, and in which family groupings may change between database releases.
PRINTS was originally built as a single ASCII (text) file. To facilitate maintenance, we later developed a relational version of the resource, known as PRINTS-S (3). Here, we describe recent progress and a new development aimed at increasing the coverage of the database, notably the creation of an automatic PRINTS supplement, termed prePRINTS.
SOURCE DATABASE AND SEARCH TOOLS
PRINTS is released in major and minor versions: minor releases reflect updates, bringing the contents in line with the current version of the source database [a SWISS-PROT/TrEMBL composite (4)]; major releases denote the addition of new material to the resource. The latter are made quarterly, each release including 50 new annotated families. Four major releases have been made since the last report.
The tools available for searching PRINTS are: (i) a BLAST (5) server, for searches against sequences matched in the current version of the database (6); and (ii) the FingerPRINTScan suite (7), for searches against fingerprints contained in the current release—this affords greater specificity than the BLAST implementation (6). A recent powerful modification of FingerPRINTScan makes explicit the familial hierarchies encoded in PRINTS-S, allowing associations to be traced from sub-family to super-family relations and, where relevant, to putative distantly related clan members that share no significant sequence similarity (8).
Several other incarnations of PRINTS are also available for searching, including a Blocks-format version at the Fred Hutchinson Cancer Research Center (9), the EMOTIF database at Stanford (10), and InterPro (to which it provides a significant amount of annotation and the bulk of its hierarchical information) (11).
PrePRINTS
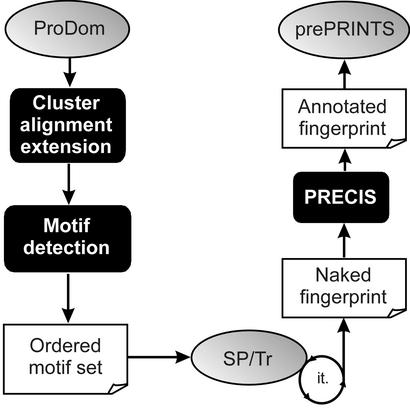
The growth of PRINTS is limited by the fact that it is maintained entirely manually, and hence it lags behind databases that are produced automatically. To begin to address this problem, we migrated the resource to a relational database management system (3). Although this facilitates routine maintenance and reduces some of the manual burdens, it does little to address database growth. We, therefore, developed an automatic supplement to PRINTS, termed prePRINTS (http://www.bioinf.man.ac.uk/prePRINTS/). This exploits an automatic pipeline (Fig. 1), which uses as input protein family clusters from ProDom (12). Motifs are detected automatically using a suite of programs, including DIALIGN (13) and CLUSTALW (14), and are used to search a SWISS-PROT/TrEMBL composite database in an iterative fashion. Naked fingerprints generated by this process are then annotated automatically using PRECIS [Protein Reports Engineered from Concise Information in SWISS-PROT (15) http://www.bioinf.man.ac.uk/cgi-bin/dbbrowser/precis/precis.cgi]. Finally, annotated fingerprints are deposited into a relational database (see Fig. 2).
Figure 1.
Illustration of the automated prePRINTS pipeline.
Figure 2.
Exerpt from a typical prePRINTS entry (for convenience, the initial and final motifs, and the matched sequence identifiers and accession numbers have been omitted). The example illustrates the fingerprint for apolipoprotein A-I, for which 4 motifs were generated by the prePRINTS pipeline. The fingerprint is ‘clean’, matching 18 sequences completely, with no partial matches, as indicated by the Summary Information. Cross-references have been generated to 6 databases, and 6 literature references have been included, the last two of which relate to crystallographic and NMR structure determinations respectively. The documentation field reports on the function of the protein and its disease associations, together with its structural and familial relationships. Keywords are also provided. In addition, a brief technical description indicates some of the parameters used to create the fingerprint. Except for the Summary Information, which was generated by the fingerprinting process, the rest of the information shown here was created fully automatically by PRECIS.
The pipeline generates 30–50 fingerprints per 24 h running on a single-processor desktop PC. The rate of conversion of these fingerprints into entries of sufficient quality for prePRINTS is ∼25% across all ProDom clusters, potentially yielding 800–900 entries/quarter — the actual rate is slower, as some human validation is necessary, for example, to discard non-specific ‘noisy’ motifs, or to eliminate restrictive motifs (i.e. those not found in all family members). The rest of the system is largely automated, so there is likely to be some redundancy with PRINTS. Nevertheless, prePRINTS serves as a valuable PRINTS ‘incubator’, wherein entries are manually refined before accession to PRINTS itself. PrePRINTS 1.0 contains 250 entries.
AVAILABILITY
For local installation, PRINTS flat-files may be retrieved from the anonymous-ftp servers at Manchester (ftp://ftp.bioinf.man.ac.uk/pub/prints), HGMP-RC (ftp://ftp.hgmp.mrc.ac.uk/pub/database/prints), EBI (ftp://ftp.ebi.ac.uk/pub/databases), EMBL (ftp://ftp.embl-heidelberg.de) and NCBI (ftp://ncbi.nlm.nih.gov). prePRINTS is available from the Manchester server.
CONCLUSION
A limitation in using protein family databases to infer function of newly-determined sequences is that of coverage; clearly, the diagnostic capability of a database is restricted to the entries it contains. The growth of PRINTS has been restricted by its manual maintenance, causing it to lag behind largely automatically-generated counterparts, such as Pfam (16). However, prePRINTS will help to increase the family coverage of PRINTS, thereby improving its effectiveness as a tool for protein sequence analysis and genome annotation.
Acknowledgments
ACKNOWLEDGEMENTS
PRINTS is built and maintained at the University of Manchester with support from the Royal Society (T.K.A. is a Royal Society University Research Fellow) and the Centre for Integrative Genomic Medical Research (A.U.). We are grateful for individual support from the MRC (A.G.), the BBSRC (A.M.), the EPSRC (N.M.), the EC (P.B.) and BioFocus (G.M.).
REFERENCES
- 1.Attwood T.K. (2001) A compendium of specific motifs for diagnosing GPCR subtypes. Trends Pharmacological Sci., 22, 162–165. [DOI] [PubMed] [Google Scholar]
- 2.Attwood T.K., Beck,M.E., Bleasby,A.J. and Parry-Smith,D.J. (1994) PRINTS — A database of protein motif fingerprints. Nucleic Acids Res., 22, 3590–3596. [PMC free article] [PubMed] [Google Scholar]
- 3.Attwood T.K., Croning,M.D.R., Flower,D.R., Lewis,A.P., Mabey,J.E., Scordis,P., Selley,J. and Wright,W. (2000) PRINTS-S: the database formerly known as PRINTS. Nucleic Acids Res., 28, 225–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 6.Wright W., Scordis,P. and Attwood,T.K. (1999) BLAST PRINTS — an alternative perspective on sequence similarity. Bioinformatics, 15, 523–524. [DOI] [PubMed] [Google Scholar]
- 7.Scordis P., Flower,D.R. and Attwood,T.K. (1999) FingerPRINTScan: intelligent searching of the PRINTS motif database. Bioinformatics, 15, 799–806. [DOI] [PubMed] [Google Scholar]
- 8.Attwood T.K., Blythe,M.J., Flower,D.R., Gaulton,A., Mabey,J.E., Maudling,N., McGregor,L., Mitchell,A.L., Moulton,G., Paine,K. and Scordis,P. (2002) PRINTS and PRINTS-S shed light on protein ancestry. Nucleic Acids Res., 30, 239–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henikoff J., Greene,E.A., Pietrokovski,S. and Henikoff,S. (2000) Increased coverage of protein families with the Blocks Database servers. Nucleic Acids Res., 28, 228–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang J.Y. and Brutlag,D.L. (2001) The EMOTIF database. Nucleic Acids Res., 29, 202–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apweiler R., Attwood,T.K., Bairoch,A., Bateman,A., Birney,E., Biswas,M., Bucher,P., Cerutti,L., Corpet,F., Croning,M.D.R., Durbin,R., Falquet,L., Fleischmann,W., Gouzy,J., Hermjakob,H., Hulo,N., Jonassen,I., Kahn,D., Kanapin,A., Karavidopoulou,Y., Lopez,R., Marx,B., Mulder,N.J., Oinn,T.M., Pagni,M., Servant,F., Sigrist,C.J.A. and Zdobnov,E.M. (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res., 29, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corpet F., Servant,F., Gouzy,J. and Kahn,D. (2000) ProDom and ProDom-CG: tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res., 28, 267–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgenstern B. (1999) DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics, 15, 211–218. [DOI] [PubMed] [Google Scholar]
- 14.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reich J.R., Mitchell,A., Goble,C.A. and Attwood,T.K. (2001) PRECIS: Protein Reports Engineered from Concise Information in SWISS-PROT. IEEE Intelligent Systems, 16, 42–51. [DOI] [PubMed] [Google Scholar]
- 16.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]