Abstract
We have established the INFEVERS — INternet periodic FEVERS — website (which is freely accessible at http://fmf.igh.cnrs.fr/infevers/). Our objectives were to develop a specialist site to gather updated information on mutations responsible for hereditary inflammatory disorders: i.e. Familial Mediterranean Fever (FMF), TRAPS (TNF Receptor 1A Associated Syndrome), HIDS (HyperIgD Syndrome), MWS (Muckle-Wells Syndrome)/FCU (Familial Cold Urticaria)/CINCA (Chronic Infantile Neurological Cutaneous and Articular Syndrome). Contributors submit their novel mutations through a 3 step form. Depending on the disease concerned, a member of the editorial board is automatically solicited to overview and validate new submissions, via a special secured web interface. If accepted, the new mutation is available on the INFEVERS web site and the discoverer, who is informed by email, is credited by having his/her name and date of the discovery on the site. The INFEVERS gateway provides researchers and clinicians with a common access location for information on similar diseases, allowing a rapid overview of the corresponding genetic defects at a glance. Furthermore, it is interactive and extendable according to the latest genes discovered.
INTRODUCTION
Hereditary inflammatory disorders are group of diseases characterized by self-resolving attacks of fever accompanied by somewhat specific clinical features [(1,2) and references herein]. While under-recognized for years, these disorders have been extensively characterized recently, with four genes, discovered within the last five years, accounting for seven OMIM diseases. As such, molecular screening has became the tool of choice in assisting clinicians with diagnosis and it is not uncommon that more than one gene per patient is tested. Unfortunately, a large amount of mutations still remain unidentified. Our objectives were to develop a specialist site to gather, in a single place, updated information on genetic defects responsible for all hereditary inflammatory disorders. The entries for the four genes discovered so far are already functional and are managed by the following editorial board (Table 1).
Table 1. The four entries of the INFEVERS website.
| Gene (in order of discovery) | Disease(s) (Ref) | OMIM | Editor |
|---|---|---|---|
| MEFV (MEditerranean FeVer) | FMF (4,5) | 249 100 | Dr I Touitou, Montpellier, France |
| TNFRSF1A (Tumor Necrosis Factor Receptor 1A) | TRAPS (6) | 142 680/134 610 | Dr C Dodé, Paris, France Dr M McDermott, London, UK |
| MVK (Mevalonate Kinase) | HIDS (7,8) | 260 920 | Dr L Cuisset, Paris, France |
| CIAS1 (Cold Induced Autoinflammatory Syndrome 1) | MWS (9)/FCU (9)/CINCA (10) | 191 900/120 100/607 115 | Dr H Hoffman, La Jolla, USA |
SITE PRESENTATION
The Familial Mediterranean Fever (FMF) website (http://fmf.igh.cnrs.fr) is a thematic grouping of three distinct but interconnected projects, accessible by WWW: MetaFMF database, the INFEVERS database and the FMF2002 (September 23–27, 2002—La Grande-Motte, France) congress site. The goal of the INFEVERS web site is to provide a genetic database for all hereditary inflammatory disorders.
The main development of the INFEVERS site is focused on the mutation data submission. Identified contributors are allowed to enter data in a secure and easy to use interface.
The FMF entry is linked to the metaFMF database. Validated FMF INFEVERS mutations are the source of the scrolling lists used in the 2nd step of the metaFMF database submission module (3).
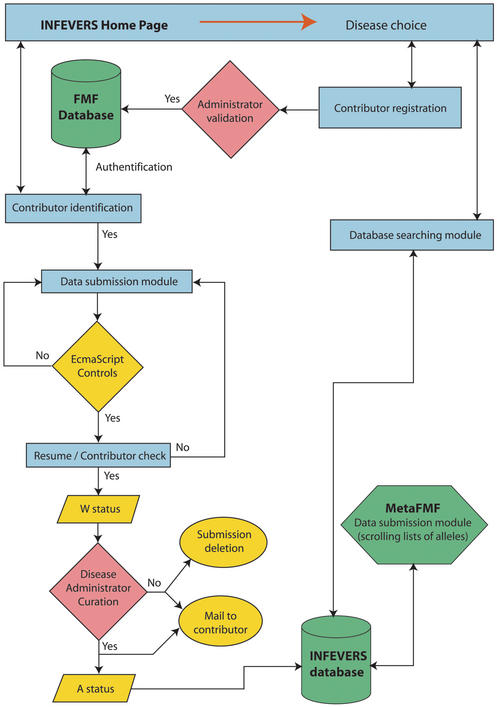
As shown in the data pathway (Fig. 1), the INFEVERS environment contains several automatic and manual controls.
Figure 1.
Overview of the INFEVERS data pathway. (i) blue rectangles: contributor interactions, (ii) yellow shapes: automatic procedures and (iii) red diamonds: administrator controls.
Extensive information on a specific disease/mutation can be easily retrieved through a search module.
CONTRIBUTOR SUBSCRIPTION
Data submission is only accessible to authenticated researchers, i.e. who are registered in the interconnected MetaFMF/INFEVERS databases. The contributor has a special form for this purpose, which contains the following criteria of identification: name, address, email, password, URL. The MetaFMF and INFEVERS subscription modules have a quite different treatment procedure. Upon confirmation by the administrator(s), an identification code is sent to the contributor. Then, he/she can enter his/her identification code and password to be authorized to submit data into the INFEVERS database.
DATA SUBMISSION PROCESS
To submit a new mutation, the identified contributor first needs to select the disease among those available in the INFEVERS database. The accession form will then display 3 successive steps:
Step 1: definition of the mutation (name, location, alteration,…)
Step 2: consequences and related data concerning this mutation (number of control chromosomes tested, disease-related symptoms, ancestry,…)
Step 3: authorship concerning the discoverer(s) (name, type, ID and URL of the publication,…)
Most of criteria choices are radio-buttons, checkboxes, or scrolling lists, to limit lacking data. A few text zones are also available for additional information. Prior to sending data to the disease administrator, the contributor has the possibility to re-access previous steps from a summary page, in order to modify or complete submitted data.
There should be no need for data to be updated or modified, thus, if a contributor would like to change some parameters of his/her former submission, he/she currently must request to do so by email to the disease administrator.
SUBMISSION AND DATA QUALITY CONTROL SYSTEMS
Many different control systems have been implemented to ensure INFEVERS data coherence (see Fig. 1). Firstly, the WWW data submission form works under a large number of instantaneous quality controls developed in ECMAscript language in order to detect mistakes, missing data and incoherent choices. For example, (i) if a mutation is known to change or define a RFLP, the contributor must choose a restriction enzyme from the list, (ii) if the gene's location is defined as an exon, but the number of the intron is filled in instead of the exon number, a warning message will be displayed and so on. Secondly, all submitted mutation data for a given disease are checked by the corresponding administrator(s) before definitive validation. In this way, different status of data validation are possible. The new data waiting for the administrator(s) approval are labelled W (Waiting), whereas the definitively validated data are labelled A (Accepted). Only the data in status ‘A’ will be displayed in the search module.
After each new submission, the INFEVERS administrator(s) automatically receive(s) an email. Within an intranet data management site, he/her will access all the mutation information concerning the disease he/she is in charge of. From there, he/she can check the new entries and decide to validate or not the data. Furthermore, it is possible to modify an already validated mutation from this intranet.
Whatever the administrator decides, contributors receive an automatically generated email with an indication of the administrator's decision.
Firstly, this system prevents data corruption within the database, and unauthorised data modification. Secondly, in the case of FMF disease, as the INFEVERS mutation entries represent the different values of the scrolling list in the MetaFMF data submission form, this safe data pathway avoids incoherence in allele names between the two web sites.
DATA SEARCHING AND RETRIEVAL
All WWW users have access to search the entire database module, disease by disease. The first connection will display all the mutations (accepted by the administrator) for the selected disease in a multi-criterion table. To refine the search in order to compare the different entries in term of location, alteration, technique(s) used, consequences, etc, the procedure is quite simple: Researchers just have to click on the parameter of interest to add it to the search filter. He/she can extend this filter as far as he/she wants.
DESIGN AND IMPLEMENTATION
The mutation data and administrator's parameters are managed using the MySQL relational database server. MySQL is an Open Source Database under the GNU General Public License (GPL).
The dynamic web pages were implemented with ECMAscript (http://www.ecma.ch) within the HTML code in order to perform controls on submission.
The correlation between the HTML page and the database is driven with Perl/CGI scripts (http://www.perl.com). The web site is running under an Apache HTTP server (http://httpd.apache.org).
As our first wish was to create a worldwide tool, nearly all currently available web browsers, both old and current versions, from various OS (Operating System) were tested in the contributor subscription and the data submission processes.
An efficient way to upgrade a web site and a database structure is to minimize downtime. The developers team, therefore, uses an alternate (development) mirror site independent from the open one. After data coherence and bug free controls the working draft is switched onto the public W3 server.
DISCUSSION AND FURTHER DEVELOPMENTS
The milestone of the first International Conference on FMF (September 1997, Jerusalem, Israel) was the identification of the first gene (MEFV) responsible for a periodic disease (FMF). During the second International Conference on FMF (May 2000, Antalya, Turkey), the discovery of two other genes (TNFR1A for TRAPS and MWK for HIDS) was reported. This prompted the current and future organisers to extend the title of the Conference to all hereditary inflammatory disorders. The third International Conference on FMF and Hereditary and Inflammatory disorders, to be held in September 2002, La Grande-Motte, France, will welcome the CIAS1 gene, recently pinpointed as surprisingly accounting for three different clinical entities: MWS/FCU/CINCA. The growing identification of genes responsible for hereditary inflammatory disorders means that the number of mutations to be tested and recorded is growing exponentially. As with all hereditary diseases, while the initial causal mutations are easily available from published sources, rarer mutations are often left unreported. Thanks to INFEVERS, this problem will be circumvented since from now on, discoverers are encouraged to submit mutations immediately upon identification: their ownership is protected by the real-time release of author details on the Internet. The fact that each entry is edited by an expert on the corresponding disease(s) guarantees the site to be accurate and up to date. Moreover, the site is designed so as to be extendable to any forthcoming related gene. In conclusion, we have developed INFEVERS, a website related database which offers user-friendly, accessible and complete information to professionals involved in the field of hereditary inflammatory disorders. Till now, 10 researchers/centers participated in submission information, and more than 200 professionals have visited the site in August 2002.
CITING INFEVERS
If you use the INFEVERS database as a tool for your published research, please cite this paper as reference.
REFERENCES
- 1.McDermott M.F. and Frenkel,J. (2001) Hereditary periodic fever syndromes. Neth. J. Med., 59, 118–125. [DOI] [PubMed] [Google Scholar]
- 2.Drenth J.P. and van der Meer,J.W. (2001) Hereditary periodic fever. N. Engl. J. Med., 345, 1748–1757. [DOI] [PubMed] [Google Scholar]
- 3.Puguère D., Ruiz,M., Sarrauste de Menthière,C., Masdona,B., Demaille,J. and Touitu,I. (2003) The MetaFMF website: a high quality tool for meta-analysis of FMF. Nucleic Acids Res., 31, 286–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The French FMF Consortium (1997) A candidate gene for familial Mediterranean fever. Nature Genet., 17, 25–31. [DOI] [PubMed] [Google Scholar]
- 5.The International FMF Consortium (1997) Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. Cell, 90, 797–807. [DOI] [PubMed] [Google Scholar]
- 6.McDermott M.F., Aksentijevich,I., Galon,J., McDermott,E.M., Ogunkolade,B.W., Centola,M., Mansfield,E., Gadina,M., Karenko,L., Pettersson,T. et al. (1999) Germline mutations in the extracellular domains of the 55 kDa TNF receptor, TNFR1, define a family of dominantly inherited autoinflammatory syndromes. Cell, 97, 133–144. [DOI] [PubMed] [Google Scholar]
- 7.Drenth J.P., Cuisset,L., Grateau,G., Vasseur,C., van de Velde-Visser,S.D., de Jong,J.G., Beckmann,J.S., van der Meer,J.W. and Delpech,M. (1999) Mutations in the gene encoding mevalonate kinase cause hyper-IgD and periodic fever syndrome. International Hyper-IgD Study Group. Nature Genet., 22, 178–181. [DOI] [PubMed] [Google Scholar]
- 8.Houten S.M., Kuis,W., Duran,M., de Koning,T.J., van Royen-Kerkhof,A., Romeijn,G.J., Frenkel,J., Dorland,L., de Barse,M.M., Huijbers,W.A. et al. (1999) Mutations in MVK, encoding mevalonate kinase, cause hyperimmunoglobulinaemia D and periodic fever syndrome. Nature Genet., 22, 175–177. [DOI] [PubMed] [Google Scholar]
- 9.Hoffman H.M., Mueller,J.L., Broide,D.H., Wanderer,A.A. and Kolodner,R.D. (2001) Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nature Genet., 29, 301–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feldmann J., Prieur,A.M., Quartier,P., Berquin,P., Cortis,E., Teillac-Hamel,D. and Fischer,A. (2002) Chronic infantile neurological cutaneous and articular syndrome is caused by mutations in CIAS1, a gene highly expressed in polymorphonuclear cells and chondrocytes. Am. J. Hum. Genet., 71, 198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]