Abstract
The Biodegradative Strain Database (BSD) is a freely-accessible, web-based database providing detailed information on degradative bacteria and the hazardous substances that they degrade, including corresponding literature citations, relevant patents and links to additional web-based biological and chemical data. The BSD (http://bsd.cme.msu.edu) is being developed within the phylogenetic framework of the Ribosomal Database Project II (RDPII: http://rdp.cme.msu.edu/html) to provide a biological complement to the chemical and degradative pathway data of the University of Minnesota Biocatalysis/Biodegradation Database (UM-BBD: http://umbbd.ahc.umn.edu). Data is accessible through a series of strain, chemical and reference lists or by keyword search. The web site also includes on-line data submission and user survey forms to solicit user contributions and suggestions. The current release contains information on over 250 degradative bacterial strains and 150 hazardous substances. The transformation of xenobiotics and other environmentally toxic compounds by microorganisms is central to strategies for biocatalysis and the bioremediation of contaminated environments. However, practical, comprehensive, strain-level information on biocatalytic/biodegradative microbes is not readily available and is often difficult to compile. Similarly, for any given environmental contaminant, there is no single resource that can provide comparative information on the array of identified microbes capable of degrading the chemical. A web site that consolidates and cross-references strain, chemical and reference data related to biocatalysis, biotransformation, biodegradation and bioremediation would be an invaluable tool for academic and industrial researchers and environmental engineers.
INTRODUCTION
A project was recently undertaken in our laboratory to describe the phylogenetic distribution of known microorganisms capable of degrading hazardous substances. The goals of this analysis were to search for patterns of degradative processes within a phylogenetic context, to gain insights into the evolution of those processes and to aid in the design of genetic probes for environmental detection of degradative microbes. The preliminary analysis resulted in the identification of over 500 bacteria that degraded or transformed a total of 138 environmentally important chemicals and showed that known degradative bacteria are not, in fact, evenly distributed evolutionarily (1). This investigation and the identification of evolutionary patterns, however, was seriously hindered by strain data that was difficult to locate and compile and was often not comparable between strains. Similarly, for any given environmental contaminant, there existed few resources that could provide comparative information on the array of identified microbes capable of degrading the chemical. The Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ) does provide a list, organized by chemical degraded, of degradative strains in its collection (http://www.dsmz.de/strains/degradtn.htm) but this list is limited to strains in the DSMZ collection and to data displayed on their strain accession pages. The difficulties encountered in this study underscored the potential usefulness of a database that would consolidate strain-level, microbial data on biodegradative/biotransforming microorganisms. A single, comprehensive, easily accessible source of information on biodegradative microorganisms and the compounds they degrade, organized within a phylogenetic framework, would be an invaluable resource for bioremediation research, providing practical information on which microbes degrade hazardous substances; the availability of, growth conditions for, and the degradative processes associated with those strains, and links to additional data located in related databases.
To address these deficiencies and provide such a resource, we have developed the Biodegradative Strain Database (BSD: http://bsd.cme.msu.edu) curated at the Center for Microbial Ecology at Michigan State University. The goals of the BSD are to:
consolidate and provide rapid access to comparative data on known biodegradative microorganisms and the hazardous substances they degrade as a freely available resource for researchers and field practitioners;
facilitate comparative analyses and highlight deficiencies in our current knowledge base;
provide corresponding microbiological data to complement and integrate with the chemical and metabolic data of the University of Minnesota Biocatalysis/Biodegradation Database (UM-BBD: http://umbbd.ahc.umn.edu) (2) and the phylogenetic data of the Ribosomal Database Project II (RDP II: http://rdp.cme.msu.edu/html) (3);
provide practical, microbial information that can be combined with individual site geochemical and activity information for application to contaminated-site remediation.
DATABASE DESCRIPTION
Contents and data organization
Biodegradative strains, chemical substrates and associated data are collected primarily from the scientific literature, on-line databases and from user contributions (see below). All entries in the BSD are required to have three cross-referenced data fields (core data): (1) the degradative strain, (2) a substrate (chemical) that it degrades, and (3) the reference stating that the strain degrades or transforms that substrate. Additional strain and substrate information is provided when available, including links to related databases whenever possible. The current version of the BSD (v. 2.02) contains data on 250 bacterial strains and 151 substrates with 203 supporting literature citations. Although there are eukaryotic, biodegradative microorganisms, for purposes of manageability, we are restricting the initial database to the prokaryotes.
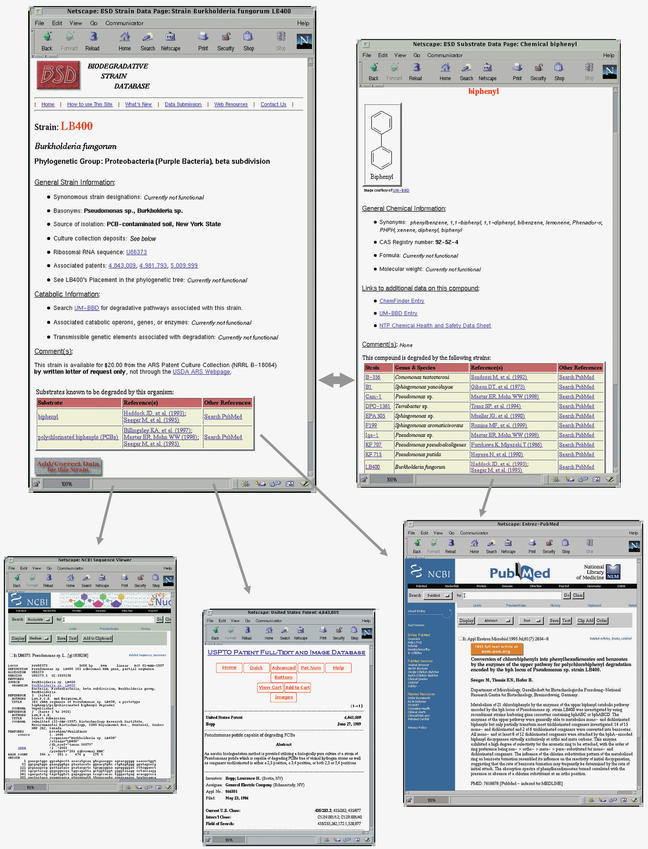
Each strain in the BSD has an individual ‘Strain Data Page’ (Fig. 1) with the list of substrates it degrades (and associated references), all other strain-level data associated with that isolate and relevant links. Similarly, each substrate in the BSD has an individual ‘Substrate Data Page’ (Fig. 1) with a list of strains that degrade or transform it, other chemical data and relevant links. These two types of individual data pages are linked to each other through their respective strain or substrate lists. References do not have individual pages in the database. Instead, citations are listed on the individual strain and chemical pages—whenever possible as hypertext links to PubMed (4) abstracts. Additional strain-associated and substrate-associated data fields currently included in or planned for the BSD are listed in Table 1.
Figure 1.
Example of a BSD individual Strain Data Page with illustration of links to core and selected ancillary data.
Table 1. Data fields included in the BSDa.
| Strain data | Substrate data |
|---|---|
| Substrates it degradesb,c | Strains that degrade itb,c |
| Genus/speciesb | Structure imagea,c |
| Taxonomic basonymsb | CAS Reg. #b |
| Phylogenetic affiliationb,c | Formulaa |
| Strain synonyms | Molecular weight |
| Source of isolationb | Synonymsb |
| Culture collection accessionsb,c | Links to UM-BBD chemical pagesb,c |
| Species-level phenotypic descriptions | UM-BBD degradative pathway links |
| Culture collection accessionsb,c | Links to ChemFinderb,c |
| rRNA GenBank accessionsb,c | Links to health and safety datab,c |
| Links to RDP datab,c | Links to vendors |
| Pertinent patentsb,c | Referencesb,c |
| Links to genomic data | Commentsb,c |
| Pathogenicity/public health datac | |
| UM-BBD degradative pathway linksc | |
| Transmissible genetic elementsc | |
| Identified gene probesc | |
| Antibiotic susceptibility | |
| Pertinent researchers/laboratories | |
| Field application examplesc | |
| Referencesb,c | |
| Commentsb,c |
aData fields not currently functional are often included in the ‘Comments’ sections.
bCurrently functional.
cProvided as links to additional data.
While the BSD is organismally oriented, the database also contains useful chemical data that can be used as an entry point for the database. The BSD also links to additional chemical information located on other websites. Chemical data unique to the BSD is the list of described microorganisms that degrade or transform it.
Accessing the data
Individual data pages can be accessed by three different avenues.
- A series of strain, substrate and reference lists is as follows:
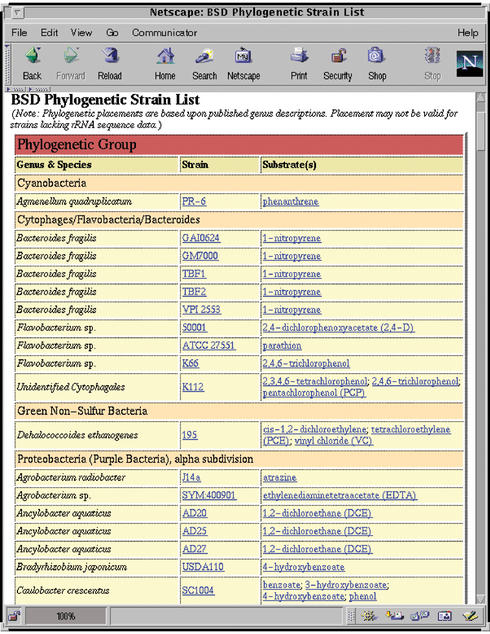
- Strain lists include the strain designation, Latin name and associated substrates for each strain. The strain designations and substrate listings serve as links to the corresponding individual data pages. The strain list can be viewed either: (1) alphabetically by strain designation, (2) alphabetically by Latin name (genus) or (3) ordered phylogenetically (Fig. 2).
- The substrate list is provided in alphabetical order with corresponding degradative strains. Again strain and substrate listings serve as links to the individual data pages.
- The reference list is alphabetical by author again with the corresponding individual strain and substrate page links. References are also provided in the table as links to PubMed (4) abstracts when available.
Keyword search: Users can search by either strain or chemical data (both core and expanded data) or citation author name. All searches, using either strain or chemical queries, result in a table of pertinent strains listed alphabetically by genus as described above with links to individual strain and chemical pages.
Phylogenetic tree interface: A phylum-level, 16S rRNA-based, phylogenetic tree is provided as a mapped image. Clicking on a branch will display a list of all BSD strains associated with that phylogenetic assemblage with links to individual strain and chemical pages.
Figure 2.
Portion of BSD phylogenetically-ordered strain list with links to individual strain and substrate data pages. The strain list is also available alphabetically by genus and strain designation to facilitate data access.
Database updates
We have divided BSD development into major and minor updates. Minor updates include incremental data expansion and corrections along with web interface updates. Major updates include the same plus improvements in functionality that require major Java programming. Our goal is to have monthly minor updates with major updates occurring two or three times per year.
BSD USER INTERACTIONS
The website contains an on-line form for the scientific community to submit new or corrected data to the BSD and a survey form to solicit user input. Data for twelve of the 250 strains in the BSD were provided by database users. Four more have been submitted for inclusion with the next update. Database users can also join the BSD mailing list.
INTERACTIONS WITH UM-BBD AND RDP II
While remaining independent, the UM-BBD (2) and the BSD have agreed to coordinate efforts where possible to better complement each other and to minimize redundancy. The UM-BBD has also offered to share technical expertise, to provide some chemical data (they currently provide chemical structures) and to establish data links between the two websites. The BSD has links from our individual substrate pages to UM-BBD chemical pages and will soon provide direct links from our individual strain and substrate pages to UM-BBD degradative pathway pages. As BSD content grows to complement UM-BBD data, reciprocal links will be established.
BSD back-end development is supported by RDP II (3) (also administrated by the Center for Microbial Ecology). In addition to GenBank (5) rRNA sequences, the BSD is now able to link to RDP II aligned rRNA sequences. BSD data will also be reorganized within the Bergey's taxonomic hierarchy, which is based upon the 16S rRNA phylogeny and is currently being implemented by the RDP II to reorganize their data.
FUTURE CHANGES AND ADDITIONS
Future additions to the BSD will include additional strains and substrates and implementation of additional data fields (Table 1). Data collection is currently focusing on microorganisms associated with metal reduction and on making the BSD more compatible with the UM-BBD. The next upgrade will include more sophisticated search engine output. All searches, using either strain or chemical queries, currently result in a strain-oriented list with links to individual data pages. The improved output will also provide search results an additional substrate-oriented table simplifying data access when searching the database by substrate related queries. Individual strain pages will also be upgraded to include species-level phenotypic descriptions and public health and pathogenicity information. A more sophisticated phylogenetic tree interface is planned and user-manipulable data tables to facilitate comparative analyses.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Drs Larry Wackett, Lynda Ellis and Doug Hershberger at the UM-BBD for their cooperation and Drs Harry Beller, Nico Boon, Nick Coleman, Alasdair Cook, Ron Crawford, Natsuko Hamamura, Minna Laine, Andrew Laurie, Thomas Moorman, Sylvie Rabot, Jorge Rodrigues, Anna Maria Solanas Canovas, Sebastian Sorensen, and G. Zaitsev, Mr James Swezey and Mr Hector Ayala for data contributions. Supported by the Great Lakes Mid-Atlantic Center for Hazardous Substance Research (GLMAC).
REFERENCES
- 1.Urbance J.W., Kukor,J.J. and Tiedje,J.M. (1999) Analysis of the phylogenetic distribution of biodegradative bacteria. Proceedings of Pseudomonas'99: Biotechnology Pathogenesis. Maui, HI.
- 2.Ellis L.B.M., Hershberger,C.D., Bryan,E.M. and Wackett,L.P. (2001) The University of Minnesota Biocatalysis/Biodegradation Database: Emphasizing Enzymes. Nucleic Acids Res., 29, 340–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maidak B.L., Cole,J.R., Lilburn,T.G., Parker,C.T., Saxman,P.R., Farris,R.J., Garrity,G.M., Olsen,G.J., Schmidt,T.M. and Tiedje,J.M. (2001) The RDP-II (Ribosomal Database Project). Nucleic Acids Res., 29, 173–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wheeler D.L., Church,D.M., Lash,A.E., Leipe,D.D., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Tatusova,T.A., Lukas Wagner,L. and Rapp,B.A. (2002) Database resources of the National Center for Biotechnology Information: 2002 update. Nucleic Acids Res., 30, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J., Rapp,B.A. and Wheeler,D.L. (2000) GenBank. Nucleic Acids Res., 30, 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]