Abstract
Multitype zinc-finger proteins of the Friend of GATA/U-shaped (Ush) class function as transcriptional regulators of gene expression through their modulation of GATA factor activity. To better understand intrinsic properties of these proteins, we investigated the expression and function of the ush gene during Drosophila embryogenesis. ush is dynamically expressed in the embryo, including several cell types present within the mesoderm. The gene is active in the cardiogenic mesoderm, and a loss of function results in an overproduction of both cardial and pericardial cells, indicating a requirement for the gene in the formation of these distinct cardiac cell types. Conversely, ectopic expression of ush results in a decrease in the number of cardioblasts in the heart and the inhibition of a cardial cell enhancer normally regulated by the synergistic activity of the Pannier and Tinman cardiogenic factors. These findings suggest that, similar to its known function in thoracic bristle patterning, Ush functions in the control of heart cell specification through its modulation of Pannier transcriptional activity. ush is also required for mesodermal cell migration early in embryogenesis, where it shows a genetic interaction with the Heartless fibroblast growth factor receptor gene. Taken together, these results demonstrate a critical role for the Ush transcriptional regulator in several diverse processes of mesoderm differentiation and heart formation.
GATA zinc-finger transcription factors are expressed in a variety of cell types, where they regulate gene expression during their control of cell commitment and differentiation (reviewed in refs. 1 and 2). Members of the GATA4, -5, and -6 subfamily are expressed in overlapping patterns during early heart development in the mouse, where they likely function in the establishment of the cardiac cell lineage. However, genetic studies thus far have failed to show a definitive role for any of the genes in cardiomyocyte specification. Loss of GATA4 function does result in aberrant cardiac morphogenesis, because null mice form bilateral heart tubes that fail to fuse at the ventral midline (3, 4). Nonetheless, normal specification of the cardiac cell lineage occurs because these tubes contain differentiated cardiomyocytes that express numerous contractile protein genes. Likewise, insights have not been gained from the analysis of other gene mutants as GATA5 null mice are viable (4) and GATA6 homozygotes die shortly after implantation (5, 6). Because functional redundancies may exist between GATA4, -5, and -6, proof of a requirement for these proteins in cardiomyocyte specification may occur only through the analysis of mice mutant for at least two of the genes.
Recent studies in zebrafish and Drosophila have demonstrated an essential function for two GATA factors in the early events of heart formation. The zebrafish faust (fau) gene, which encodes the GATA5 transcription factor, is required for the formation of normal numbers of myocardial precursors and the proper expression of several cardiac-expressed genes, including nkx2.5 (7). fau function is also needed for heart and endoderm morphogenesis, including the production of ventricular tissue. Forced expression of GATA5 results in the de novo activation of several myocardial genes and the ectopic production of contracting heart-like tissue. Such results demonstrate an integral role for this GATA factor in cardiac cell production, differentiation, and morphogenesis during zebrafish development.
During Drosophila embryogenesis, the GATA gene pannier (pnr; refs. 8 and 9) is expressed in the cardiogenic mesoderm, where it is required for cardial cell formation while repressing the overproduction of a pericardial cell type (10). pnr can function as a cardiac identity gene because its forced expression results in supernumerary cardial cells at the expense of other dorsal mesodermal cell lineages. Pnr works synergistically with the Tinman (Tin) homeodomain protein (11–13) in the activation of a heart enhancer for the D-mef2 differentiation gene and the specification of the cardioblast fate (10, 14, 15). Mouse GATA4 can also perform as a cardiogenic factor in the Drosophila system, suggesting an evolutionarily conserved function between Pnr and GATA4 in the early stages of heart development.
In addition to this functional synergism of Pnr and Tin, cardiac-expressed GATA proteins are known to interact with heterologous transcription factors, including NFAT3, Nkx-2.5, and Friend of GATA-2 (FOG-2; refs. 16–22). FOG-2 is a multitype zinc-finger protein that is coexpressed with the GATA factors in several cell types in the mouse, including the developing and mature heart. FOG-2 can bind to the amino-terminal zinc-finger of GATA4 and affect GATA4 transcriptional activity in its regulation of various cardiac promoters in cell culture studies (20–22). These results suggest the involvement of FOG-2 as a modulator of GATA factor function in the activation of cardiac-expressed genes during cardiomyocyte specification and subsequent heart development.
U-shaped (Ush) is a multitype zinc-finger protein of Drosophila that is structurally related to FOG-2 (23). Comparable to the interactions of GATA4 and FOG-2, Pnr and Ush heterodimerize through the amino-terminal zinc finger of Pnr (24), and genetic studies have shown that Ush antagonizes the function of Pnr in the establishment of the thoracic bristle pattern in the adult (24, 25). Given the molecular and functional similarities between the GATA and multitype zinc-finger proteins of vertebrates and flies and the recent demonstration of a cardiogenic function of pnr, we looked for a possible role for ush in Drosophila heart development. In this report, we show that ush is expressed in the embryonic mesoderm including the cardiogenic region. Genetic studies demonstrate a requirement for ush function in the normal migration of the mesoderm and in the proper formation of cardiac cells. Our results are consistent with a model in which Ush modulates the function of Pnr in heart cell specification, similar to its known function in neuronal precursor cell determination.
Materials and Methods
Fly Strains.
y w67c23 served as our wild-type stock. The ush strains used in this study were obtained from the Umea Stock Center (Umea, Sweden) and included the presumed point mutations ush1 cn1 bw1 sp1/CyO and ush2 cn1 bw1 sp1/CyO and the ush deletion Df(2L)al/CyO, ftz-lacZ. These three mutations have been classified previously as embryonic lethals and germ band retraction amorphs (26). The ush1/SM6, Roi, eve-lacZ and ush2/SM6, Roi, eve-lacZ stocks were further established to discriminate between the mutant and balancer chromosomes. The D-mef2 enhancer-lacZ gene used was IIA341 (10, 15). The Gal4 driver strain used to direct gene expression in the mesoderm was twi-Gal4 (27). The UAS-ush stock was generated in this study by standard PCR amplification of a full-length ush cDNA cloned in pBluescript (Research Genetics, Huntsville, AL), by using oligonucleotides (Sigma–Genosys) to engineer 5′ XhoI (pUASTush1: ATATACTCGAGGAATAATTGTATTACAGTGAGTAGTGAG) and 3′ XbaI (pUASTush2: AAATATCTAGACTCCGGGAATACTGTCGTCGATTG) restriction sites at the ends of the 4.5-kb cDNA insert. This fragment was then cloned into the corresponding sites of the pUAST transformation vector (28), with insert orientation and integrity confirmed by DNA sequencing. The pUAST-ush DNA was injected into y w67c23 embryos, and multiple transgenic lines were established by using standard transformation procedures (14). Eight lines were tested and showed a moderate to strong dorsal vessel phenotype. The heartless (htl) allele used in this study was htlAB42/TM3, Sb, ftz-lacZ, characterized as a null allele based on molecular and phenotypic analyses (29).
RNA in Situ Hybridization and Immunohistochemical Staining of Embryos.
DNAs containing ush or pnr cDNAs cloned in pBluescript were linearized by restriction digestion and used as templates to generate antisense cRNA probes by using the DIG RNA labeling kit (Roche Molecular Biochemicals). RNA in situ hybridization and sectioning of embryos were performed as described (10). The same study also describes the methods used for immunostaining of embryos with mouse anti-β-galactosidase, rabbit anti-Drosophila myocyte enhancer factor 2 (D-MEF2), and mouse anti-Even-skipped (Eve) antibodies.
Gene Expression Analyses in Mutant and Gal4/UAS Embryos.
Embryos obtained from the mating of twi-Gal4 and UAS-ush flies were collected at 29°C, whereas all other embryos were collected at 23°C. To analyze the function of the D-mef2 cardial cell enhancer in ush mutant embryos, ush1/SM6, Roi, eve-lacZ or Df(2L)al/CyO, ftz-lacZ males were crossed to y w; IIA341–44 females. Non-Cy F1 virgins of the genotype ush1/+;IIA341–44/+ or Df(2L)al/+;IIA341–44/+ were then backcrossed to males of the respective parental strains. Expression of the D-mef2-lacZ fusion gene was assayed by staining for β-galactosidase activity. Homozygous embryos were identified by the failure of germ band retraction, and the number of cardial cells present per hemisegment was averaged over 15 mutant embryos.
ush embryos were also stained for D-MEF2 and β-galactosidase protein to determine whether there was an increase in the number of cardial cell nuclei. Homozygous embryos were identified by the germ band retraction phenotype and the absence of the β-galactosidase marker associated with the balancer chromosome. Nuclei counts were obtained from an average of 10 wild-type or ush mutant embryos. To determine the number of Eve-expressing cells, embryos were collected from the mating of ush stocks and stained for Eve and β-galactosidase with homozygous embryos identified by the absence of the marker protein. The average number of Eve-expressing pericardial cells was determined from the count of 20 mutant embryos. To analyze the activity of cardial cell markers in embryos with forced Ush expression, UAS-ush males were crossed to twi-Gal4 or twi-Gal4;IIA341–44 females, with the resulting embryos stained with anti-D-MEF2 or anti-β-galactosidase antibodies, respectively.
Expression Vectors, Cell Transfections, and CAT Assays.
Drosophila S2 cells were grown in M-3 medium (Quality Biologicals, Gaithersburg, MD) supplemented with 10% FBS. Cells (1 × 105 per transfection) were cotransfected with the indicated expression vectors (2 μg per transfection) and the D-mef2-CAT reporter plasmid (1 μg per transfection) by using the calcium phosphate precipitation method. Normalization of transfection efficiency with β-galactosidase activity and CAT assays with a CAT ELISA kit (Roche Molecular Biochemicals) were performed as described (30). For the construction of expression vectors, full-length cDNAs for pnr, ush, and tin were subcloned into the pIZ/V5-His plasmid (Invitrogen). For the Tin (N-Q) construct, the coding region for the homeodomain in the wild-type Tin expression vector was replaced by a DNA fragment from the A6-NK4HD(NQ) plasmid (31) containing the mutated sequence corresponding to an Asn to Gln substitution at amino acid position 351. The D-mef2-CAT reporter plasmid was constructed by inserting the DNA fragment from the D-mef2–341 vector (15) into the BamHI site of the pBLCAT2 plasmid (Promega).
Analysis of a Mesodermal Cell Migration Phenotype.
Embryos were collected from the independent matings of the ush1/SM6, Roi, eve-lacZ and htlAB42/TM3, Sb, ftz-lacZ stocks and from an intercross of these flies to produce ush1/+;htlAB42/+ transheterozygotes. Embryo populations were stained for D-MEF2 and β-galactosidase proteins. Homozygous or transheterozygous mutant embryos were identified by the absence of β-galactosidase activity associated with the presence of the balancer chromosome. The migration of mesodermal cells marked by D-MEF2 expression was analyzed in embryo cross sections as described (10).
Results
ush Expression in the Embryonic Mesoderm.
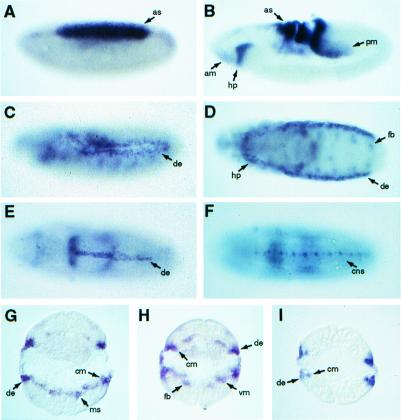
The ush gene exhibits a dynamic pattern of expression during embryogenesis. Gene transcripts are first detected at high levels in the primordium of the amnioserosa at stage 5 (Fig. 1A). Additional expression is observed in germ band extending embryos in cells of the developing anterior and posterior midgut and in hemocyte precursors present in the cephalic mesoderm (Fig. 1B). By stage 11, ush RNA is detected in the dorsal ectoderm and in precursor cells of the hemocytes and fat body (Fig. 1 C and D). By late embryogenesis, ush expression is greatly diminished, but transcripts are still observed in the dorsal ectoderm during dorsal closure (Fig. 1E) and cells within, or associated with, the central nervous system (Fig. 1F). To investigate the possible expression of ush in mesodermal cells underlying the dorsal ectoderm, we examined cross sections of embryos at stage 11. ush RNA is detected in a changing pattern in this germ layer, initially throughout most of the mesoderm (Fig. 1G) and then in subpopulations of cells, including precursors of the fat body, visceral mesoderm, and cardiogenic mesoderm (Fig. 1H). As a comparison, the presence of pnr RNA in the dorsal ectoderm and cardiogenic mesoderm is also shown (Fig. 1I; ref. 10). Therefore, ush is expressed in the dorsal mesoderm, where it could function in the early stages of heart formation.
Figure 1.
ush gene expression during Drosophila embryogenesis. (A–C) Lateral views of ush RNA in stage 5, 8, and 11 whole-mount embryos. (D and F) Ventral views of ush RNA in stage 11 and 16 whole-mount embryos. (E) Dorsal view of ush RNA in a stage 16 whole-mount embryo. (G and H) Cross sections of stage 11 embryos stained for ush RNA. (I) Cross section of a stage 11 embryo stained for pnr RNA. Abbreviations: am, anterior midgut primordium; as, amnioserosa primordium; cm, cardiogenic mesoderm; cns, central nervous system; de, dorsal ectoderm; fb, fat body precursors; hp, hemocyte primordium; ms, mesoderm; pm, posterior midgut primordium; vm, visceral mesoderm precursors.
ush Function Is Required for Proper Cardiac Cell Specification.
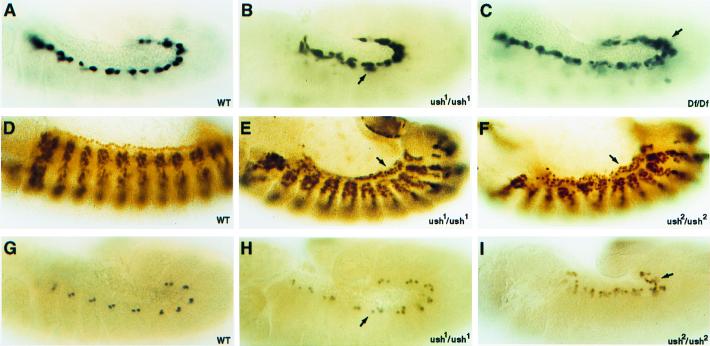
pnr mutant embryos show a loss of contractile cardial cells and an overproduction of certain nonmuscle pericardial cells in the heart-forming region (10). To identify a possible role for ush in these cardiogenic processes, we looked for alterations in cardiac cell production in mutant embryos. The ush alleles used in this analysis were ush1 and ush2, believed to be hypomorphic mutations of the gene, and Df(2L)al, a chromosome deletion that represents a ush null mutation. The D-mef2 heart enhancer-lacZ fusion gene serves as a cardial cell marker as it is detected in progenitors of these cells around stage 11 and thereafter in two, then four cardioblasts per hemisegment of the forming dorsal vessel (14, 15, 32). Embryos homozygous for either a point mutation or deletion of the gene showed an increase in the number of cells expressing the reporter gene as compared with the wild-type embryo (Fig. 2A). In ush1 embryos, a few hemisegments contained up to nine positive cells with an average of six cardial cells present in many clusters (Fig. 2B). In ush-deficiency embryos, a comparable increased density of cardial cells was obtained (Fig. 2C).
Figure 2.
ush gene functions during Drosophila cardiogenesis. (A, D, and G) WT, wild-type embryos. (B, C, E, F, H, and I) Homozygous ush embryos of the indicated genotype. (A–C) Lateral views of stage 12 embryos stained for lacZ reporter gene expression under the control of a D-mef2 cardial cell enhancer. (D–F) Lateral views of stage 13 embryos stained for D-MEF2 protein in cardial cells. (G–I) Lateral views of stage 12 embryos stained for Eve protein in a subset of pericardial cells. Arrows point to the increased numbers of cardial or pericardial cells in the ush mutant embryos.
D-MEF2 protein also marks cardioblasts as it is detected in the nuclei of all cardial, but not pericardial cells of the forming dorsal vessel. In wild-type embryos at stage 13, the germ band has retracted with cardioblasts migrating dorsally, separating from the dorsal somatic muscles. A lateral view at this stage shows a single row of cells that contains six stained nuclei per hemisegment (Fig. 2D). In contrast, ush mutant embryos possess supernumerary cardioblast nuclei. ush1 (Fig. 2E) and ush2 (Fig. 2F) embryos contain up to 12 nuclei per hemisegment with eight cells per cluster observed on average. Similar results were also obtained with ush-deficiency embryos (data not shown). Therefore, reducing or completely eliminating ush function leads to an increased production of cardial cells. Intriguingly, the ush heart phenotype uncovered by the analysis of these two markers directly contrasts the absence of cardial cells observed in pnr loss-of-function embryos.
We also assayed for the production of pericardial cells in ush mutant embryos, using Eve protein as a marker for a subset of these cells. In wild-type embryos at stage 12, there exist 11 Eve-positive clusters within the dorsal mesoderm, each containing about three cells (Fig. 2G). In contrast, the number of Eve-expressing pericardial cells increased in homozygous ush1 and ush2 embryos to an average of 5–6 per cluster (Fig. 2 H and I). A similar increase in pericardial cell number was also observed in homozygous Df(2L)al embryos (data not shown). Thus, ush gene activity is required to prevent the overproduction of this pericardial cell type, a function that has also been ascribed to the pnr gene.
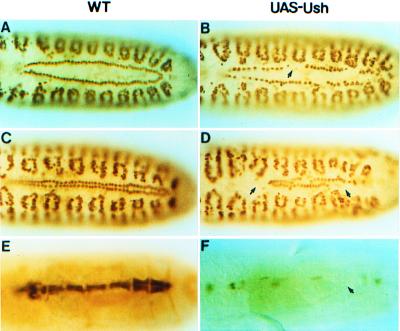
Forced Expression of ush Results in a Diminution of Cardial Cells.
Because the loss of ush function resulted in a supernumerary cardial cell phenotype, we wanted to monitor the effect of expressing the gene throughout the mesoderm using the Gal4/UAS binary system (28). D-MEF2 was used to assess the status of cardial cells, with two contiguous rows of 52 cells present in the forming or mature dorsal vessel of wild-type embryos (Fig. 3 A and C). In comparably staged embryos expressing ush throughout the mesoderm, a significant reduction in cardial cells was observed. In the two representative embryos shown, a 37% (Fig. 3B) and 50% (Fig. 3D) decrease occurred with small gaps or large deletions present within the heart tubes. The D-mef2 heart enhancer-lacZ fusion gene was used as a second marker for the cardial cells and also to assay the effect of ush expression on enhancer activity. In wild-type embryos at stage 16, the enhancer is active in eight cardial cells in most segments of the dorsal vessel (Fig. 3E). In contrast, β-galactosidase activity is greatly diminished in the hearts of ush-expressing embryos, most likely a combination of the decrease in cardial cell number and the reduced activity of the D-mef2 cardiac enhancer (Fig. 3F). Thus, forced expression of ush has a potent negative effect on cardial cell formation and enhancer function.
Figure 3.
Forced mesodermal expression of the ush gene affects heart development. (A, C, and E) WT, wild-type embryos. (B, D, and F) UAS-Ush, embryos expressing ush in the mesoderm under the control of a twi-Gal4 driver. (A–D) Expression of ush results in a reduced number of cardial cells in the forming dorsal vessel. Stage 14 (A and B) and 16 (C and D) embryos stained for D-MEF2 protein. (E and F) Expression of ush results in a decreased activity of the D-mef2 cardial cell enhancer. Stage 16 embryos were stained for β-galactosidase protein in cardial cells of the dorsal vessel. Arrows point to missing cardial cells or cardioblasts that fail to express the lacZ reporter gene.
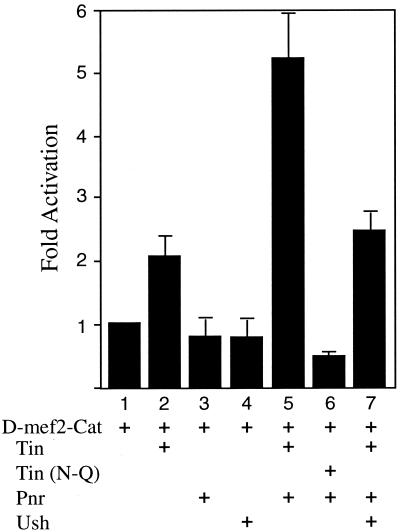
Ush Decreases the Synergistic Activation of the D-mef2 Heart Enhancer by Pnr and Tin.
It was previously shown that Pnr could function combinatorially with Tin in the regulation of the D-mef2 heart enhancer in Drosophila embryos (10). We further analyzed this synergistic activation using a transient transfection assay in cultured Drosophila cells. The activation of a CAT reporter gene linked to the D-mef2 enhancer was monitored in cells transfected independently with Pnr, Tin, and Ush or with various combinations of the factors. The expression of Tin alone activated the enhancer about 2-fold above the basal level, whereas neither Pnr nor Ush affected enhancer activity (Fig. 4, lanes 1–4). Coexpression of Pnr and Tin resulted in a synergistic activation of the enhancer to a level 5–6 times that of the basal activity (Fig. 4, lane 5), and this strong induction required the binding of Tin to the D-mef2 enhancer as a Tin DNA binding mutant, Tin (N-Q), failed to synergize with Pnr in the assay (Fig. 4, lane 6). In contrast, adding Ush as a third transfected factor significantly inhibited the synergistic activation of the enhancer by Pnr and Tin (Fig. 4, lane 7). This result demonstrates that Ush can antagonize the positive functional interaction of Pnr and Tin in the regulation of the cardial cell enhancer, which is consistent with the in vivo data shown in Fig. 3F.
Figure 4.
Ush antagonizes Pnr transcriptional synergy with Tin in the regulation of the D-mef2 cardiac enhancer. Drosophila S2 cells were cotransfected with a D-mef2-CAT reporter plasmid along with Tin, Pnr, and Ush expression plasmids, as indicated. The normalized CAT activity obtained from the transfections of a test effector(s) with the reporter plasmid was divided by the corresponding value obtained with an empty vector (pIZ/V5-His) and reporter, with the value shown as fold activation. The data show that Pnr collaborates with Tin to activate the D-mef2 enhancer, and this effect is diminished by the inclusion of Ush.
ush Embryos Exhibit a Mesodermal Cell Migration Phenotype.
As previously noted, ush mutants contain an increased number of cardial cells. However, we also noticed in about half of the embryos a disparity in the cardial cell populations, ranging from a high of 8–12 per hemisegment down to regions completely devoid of cells. This complex phenotype was observed with both ush hypomorphic (Fig. 5A) and null (Fig. 5B) alleles. This sporadic loss of cells from the dorsal-most part of the mesoderm was reminiscent of a htl mutant phenotype, where the absence of the encoded fibroblast growth factor receptor homologue results in an incomplete dorsal migration of mesodermal cells (29, 33, 34). In this event, cells fail to receive the ectodermal signal needed for their further commitment, resulting in a loss of dorsal mesodermal derivatives, including cardioblasts.
Figure 5.
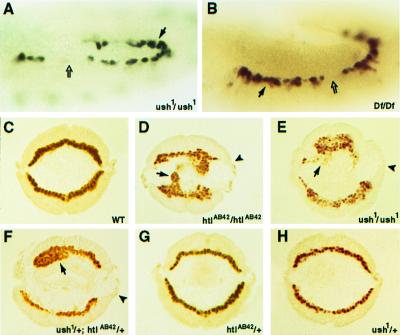
ush function in mesodermal cell migration. (A and B) Lateral views of stage 11 and 12 embryos of the genotypes ush1/ush1;IIA341–44/+ and Df(2L)al/Df(2L)al;IIA341–44/+, respectively, stained for β-galactosidase produced in cardial cells under the control of a D-mef2 heart enhancer. Solid arrows point to increased numbers of cardial cells whereas open arrows indicate the absence of cells in some hemisegments. (C–H) Analysis of mesodermal cell migration defects. Stage 10 embryos of the indicated genotypes were stained for D-MEF2 protein and sectioned to visualize the normal or abnormal spreading of the mesodermal cell layer. Arrows point to clusters of mesodermal cells, whereas arrowheads indicate the dorsal-most ectodermal cells that correspond to the normal position of cell spreading in wild-type embryos. Strong migration defects are observed in homozygous htl and ush mutants and in embryos transheterozygous for the two genes.
To determine whether the variable absence of cardial cells in ush embryos was because of a cell migration defect, we stained wild-type and mutant embryos for D-MEF2 protein present in the invaginated population of mesodermal cells. In cross sections of normal embryos at stage 10, a uniform layer is observed where the dorsal-most mesodermal cells have migrated to a position adjacent to the dorsal-most ectodermal cells (Fig. 5C). In contrast, both htl (Fig. 5D) and ush (Fig. 5E) homozygous embryos display an irregular layer where mesodermal cells remain clustered and fail to undergo their complete dorsal migration. This result suggested ush function is required for the directional migration of the mesoderm. To investigate a potential genetic interaction of htl and ush in this process, we next examined embryos that were heterozygous for mutations in each of the genes. These embryos also presented a strong mesodermal migration phenotype (Fig. 5F), suggesting the two genes function in a common genetic pathway that controls this aspect of mesoderm differentiation. As was observed with homozygous ush embryos, slightly less than half (47%; n = 46/97) of the transheterozygous embryos showed a loss of cells from the dorsal mesoderm. As a control, we likewise examined embryos individually heterozygous for htl or ush gene mutations, and such embryos exhibited a normal pattern of mesodermal cell migration (Fig. 5 G and H).
Discussion
In this report, we show that the transcriptional regulator Ush is expressed and functions during Drosophila embryogenesis. Consistent with its known requirement for germ band retraction (26), ush is abundantly expressed in the forming amnioserosa. Current results further demonstrate a function for the gene in the dorsal migration of mesodermal cells during the early development of this germ layer and the production of two cardiac cell types that contribute to the dorsal vessel. Because ush is expressed in additional cells in the embryo, the gene is likely to function in the commitment and/or differentiation of other lineages as well.
Ush is structurally similar to the vertebrate FOG proteins, and it has been shown to negatively regulate Pnr function within the larval wing disk during sensory bristle development (24, 25). When expressed alone, Pnr functions as a transcriptional activator of an enhancer for the proneural genes achaete (ac) and scute (sc), turning on these loci in the dorsocentral cluster. In cellular domains where Pnr and Ush overlap, heterodimers form between the two proteins, and these complexes serve as transcriptional repressors. Thus, Ush can antagonize the activator function of Pnr in cells where they are coexpressed, and this property suggests that Ush is functionally similar to the FOG proteins because they also modulate the activity of GATA factors in the control of gene expression (20–22, 35). Given this proven interaction of Pnr and Ush in regulating ac and sc during thoracic bristle patterning, and the recent demonstration of pnr function in cardiac cell production, we looked for ush expression in the dorsal mesoderm. Our results show that, like pnr, ush is expressed in the heart-forming region, where it could interact with pnr in regulating the production of heart precursor cells.
pnr has a dual requirement in the cardiogenic mesoderm because it is needed for the formation of cardial cells although simultaneously limiting the production of Eve-expressing pericardial cells (10). Based on these dissimilar phenotypes, we postulated Pnr might work with different combinations of factors to promote or repress the formation of cells within the distinct lineages. Recent studies have shown that Pnr and Tin act synergistically to induce cardial cells and activate gene expression, and the loss of function of either of these genes results in an absence of cardial cells. Therefore, the two work together in cardial cell specification (Fig. 6).
Figure 6.
Model of the functional interactions of Pnr, Tin, and Ush during cardial and pericardial cell specification, based on loss- and gain-of-function phenotypes. The arrow indicates a positive requirement for Tin and Pnr in cardial cell formation, whereas the solid blocked line indicates a negative role for the pnr, ush, and/or tin genetic combination in this process. The hatched blocked line indicates a negative function for these genes in the Eve-expressing pericardial cells. The model is a simplification of the total genetic complexity of cardiac cell specification in Drosophila and does not necessarily list all components required to control the production of these distinct cell types.
In contrast, Ush is a factor that normally suppresses cardial cell production. ush homozygous and hemizygous embryos show an increase in cardial cell number, and the latter finding suggests Ush control of this cell population is dose-dependent, as is the case for Ush regulation of Pnr during sensory bristle development (24). Furthermore, forced expression of Ush decreases cardial cell production and D-mef2 heart enhancer activity, whereas ectopic expression of Pnr produces extra cardial cells and expands the domain of enhancer activity. Thus, Ush displays phenotypes that are in direct opposition to those of Pnr, suggesting that it can function as an antagonist of Pnr's cardiogenic activity (Fig. 6). This conclusion is supported by the ability of Ush to inhibit the synergy of Pnr and Tin in the activation of the D-mef2 heart enhancer in cell transfection studies. As for the second cardiac phenotype, both pnr and ush are required to limit the number of Eve-expressing pericardial cells, consistent with a model in which Ush and Pnr function as corepressors in the control of these cells (Fig. 6). To summarize, our genetic studies predict that, in the wild-type embryo, pnr is expressed and functions independently of ush in precursors of the cardioblast lineage. However, in neighboring cells that include the Eve lineage precursors, the expression and function of the two most likely overlap. Future expression analyses of the two regulatory proteins at the resolution of single mesodermal cells will be required to elaborate on this genetic model in molecular terms.
About half of the ush embryos show a complex phenotype wherein certain hemisegments contain supernumerary cardial cells, whereas others are devoid of the heart precursors. Because this latter observation was reminiscent of a htl phenotype, we examined mesodermal cell migration in the mutants and also looked for a genetic interaction between ush and htl. These studies showed that ush activity is needed for the proper formation of the mesodermal monolayer, but the phenotype is only partially penetrant as the normal dorsal migration of many cells does occur. This conclusion is based on the position of the D-MEF2-expressing cells and the formation of cardiac precursors, which requires the mesodermal cells be present within the Dpp signaling domain of the dorsal ectoderm (36). Intriguingly, embryos transheterozygous for the htl and ush genes showed a strong mesodermal migration phenotype, suggesting they function in a common genetic pathway used in the movement of the cells. A possible explanation for this interaction would be that Ush functions in the activation of htl gene expression in the early mesoderm. Alternatively, it could be involved in the production of the ligand for the Htl fibroblast growth factor receptor in cells of the dorsal ectoderm, providing an attractant source for the mesodermal cells (29).
In conclusion, our current and recent results contribute to the expanding evidence for the conservation of genes controlling cardiogenesis in Drosophila and higher eukaryotes. Two common themes appear to be emerging, and such hypotheses can be further investigated both molecularly and genetically. First, it is likely that positive combinatorial interactions occur between GATA and Tin class factors in the specification of cardiac cell lineages and the activation of cardiac gene expression. Second, it is possible that antagonistic functions exist between GATA and Ush/FOG-2 type proteins in the transcription of cardiac expressed genes and in cell fate specification during early heart development. Relevant to the latter view, it will be of clear interest to compare the phenotypes of ush and fog-2 mutant embryos to identify their common or different functions in the genetic control of cardiogenesis.
Acknowledgments
We thank A. Michelson, B. Paterson, the Umea Stock Center, and the Developmental Studies Hybridoma Bank for providing genetic and molecular reagents used in this study. We also thank Karen Hensley and Rama Grenda for assistance with figures. This research was supported by grants from the Muscular Dystrophy Association and the National Institutes of Health (HL59151) (to R.A.S.). N.F. was supported by a National Institutes of Health postdoctoral training grant (T32-HD07324).
Abbreviations
- D-MEF2
Drosophila myocyte enhancer factor 2
- Eve
Even-skipped
- FOG-2
Friend of GATA-2
- htl
heartless
- Pnr
Pannier
- Tin
Tinman
- UAS
upstream activation sequence
- Ush
U-shaped
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Orkin S H. Curr Opin Genet Dev. 1996;6:597–602. doi: 10.1016/s0959-437x(96)80089-x. [DOI] [PubMed] [Google Scholar]
- 2.Parmacek M S, Leiden J M. Heart Development. San Diego: Academic; 1999. pp. 291–306. [Google Scholar]
- 3.Kuo C T, Morrisey E E, Anandappa R, Sigrist K, Lu M M, Parmacek M S, Soudais C, Leiden J M. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 4.Molkentin J D, Lin Q, Duncan S A, Olson E N. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 5.Morrisey E E, Tang Z, Sigrist K, Lu M M, Jiang F, Ip H S, Parmacek M S. Genes Dev. 1998;12:3579–3590. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. Development (Cambridge, UK) 1999;126:723–732. [PubMed] [Google Scholar]
- 7.Reiter J F, Alexander J, Rodaway A, Yelon D, Patient R, Holder N, Stainier D Y R. Genes Dev. 1999;13:2983–2995. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramain P, Heitzler P, Haenlin M, Simpson P. Development (Cambridge, UK) 1993;119:1277–1291. doi: 10.1242/dev.119.4.1277. [DOI] [PubMed] [Google Scholar]
- 9.Winick J, Abel T, Leonard M W, Michelson A M, Chardon-Loriaux I, Holmgren R A, Maniatis T, Engel J D. Development (Cambridge, UK) 1993;119:1055–1065. doi: 10.1242/dev.119.4.1055. [DOI] [PubMed] [Google Scholar]
- 10.Gajewski K, Fossett N, Molkentin J D, Schulz R A. Development (Cambridge, UK) 1999;126:5679–5688. doi: 10.1242/dev.126.24.5679. [DOI] [PubMed] [Google Scholar]
- 11.Kim Y, Nirenberg M. Proc Natl Acad Sci USA. 1989;86:7716–7720. doi: 10.1073/pnas.86.20.7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azpiazu N, Frasch M. Genes Dev. 1993;7:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- 13.Bodmer R. Development (Cambridge, UK) 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- 14.Gajewski K, Kim Y, Lee Y M, Olson E N, Schulz R A. EMBO J. 1997;16:515–522. doi: 10.1093/emboj/16.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gajewski K, Kim Y, Choi C Y, Schulz R A. Dev Genes Evol. 1998;208:382–392. doi: 10.1007/s004270050194. [DOI] [PubMed] [Google Scholar]
- 16.Durocher D, Charron F, Warren R, Schwartz R J, Nemer M. EMBO J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y, Shioi T, Kasahara H, Jobe S M, Wiese R J, Markham B, Izumo S. Mol Cell Biol. 1998;18:3120–3129. doi: 10.1128/mcb.18.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molkentin J D, Lu J, Antos C L, Markham B, Richardson J, Robbins J, Grant S R, Olson E N. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sepulveda J L, Belaguli N, Nigam V, Chen C Y, Nemer M, Schwartz R J. Mol Cell Biol. 1998;18:3405–3415. doi: 10.1128/mcb.18.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu J R, McKinsey T A, Xu H, Wang D Z, Richardson J A, Olson E N. Mol Cell Biol. 1999;19:4495–4502. doi: 10.1128/mcb.19.6.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Svensson E C, Tufts R L, Polk C E, Leiden J M. Proc Natl Acad Sci USA. 1999;96:956–961. doi: 10.1073/pnas.96.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tevosian S G, Deconinck A E, Cantor A B, Rieff H I, Fujiwara Y, Corfas G, Orkin S H. Proc Natl Acad Sci USA. 1999;96:950–955. doi: 10.1073/pnas.96.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cubadda Y, Heitzler P, Ray R P, Bourois M, Ramain P, Gelbart W, Simpson P, Haenlin M. Genes Dev. 1997;11:3083–3095. doi: 10.1101/gad.11.22.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haenlin M, Cubadda Y, Blondeau F, Heitzler P, Lutz Y, Simpson P, Ramain P. Genes Dev. 1997;11:3096–3108. doi: 10.1101/gad.11.22.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Garcia M J, Ramain P, Simpson P, Modolell J. Development (Cambridge, UK) 1999;126:3523–3532. doi: 10.1242/dev.126.16.3523. [DOI] [PubMed] [Google Scholar]
- 26.Nusslein-Volhard C, Wieschaus E, Kluding H. Roux Arch Dev Biol. 1984;193:267–282. doi: 10.1007/BF00848156. [DOI] [PubMed] [Google Scholar]
- 27.Baylies M K, Bate M. Science. 1996;272:1481–1484. doi: 10.1126/science.272.5267.1481. [DOI] [PubMed] [Google Scholar]
- 28.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 29.Gisselbrecht S, Skeath J B, Doe C Q, Michelson A M. Genes Dev. 1996;10:3003–3017. doi: 10.1101/gad.10.23.3003. [DOI] [PubMed] [Google Scholar]
- 30.Lee Y M, Park T, Schulz R A, Kim Y. J Biol Chem. 1997;272:17531–17541. doi: 10.1074/jbc.272.28.17531. [DOI] [PubMed] [Google Scholar]
- 31.Choi C Y, Lee Y M, Kim Y H, Park T, Jeon B H, Schulz R A, Kim Y. J Biol Chem. 1999;274:31543–31552. doi: 10.1074/jbc.274.44.31543. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen H T, Xu X. Dev Biol. 1998;204:550–566. doi: 10.1006/dbio.1998.9081. [DOI] [PubMed] [Google Scholar]
- 33.Beiman M, Shilo B Z, Volk T. Genes Dev. 1996;10:2993–3002. doi: 10.1101/gad.10.23.2993. [DOI] [PubMed] [Google Scholar]
- 34.Shishido E, Ono N, Kojima T, Saigo K. Development (Cambridge, UK) 1997;124:2119–2128. doi: 10.1242/dev.124.11.2119. [DOI] [PubMed] [Google Scholar]
- 35.Tsang A P, Visvader J E, Turner C A, Fujiwara Y, Yu C, Weiss M J, Crossley M, Orkin S H. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 36.Frasch M. Nature (London) 1995;374:464–467. doi: 10.1038/374464a0. [DOI] [PubMed] [Google Scholar]