Abstract
Receptor Tyrosine Kinases (RTK) are transmembrane receptors specifically found in metazoans. They represent an excellent model for studying evolution of cellular processes in metazoans because they encompass large families of modular proteins and belong to a major family of contingency generating molecules in eukaryotic cells: the protein kinases. Because tyrosine kinases have been under close scrutiny for many years in various species, they are associated with a wealth of information, mainly in mammals. Presently, most categories of RTK were identified in mammals, but in a near future other model species will be sequenced, and will bring us RTKs from other metazoan clades. Thus, collecting RTK sequences would provide a good starting point as a new model for comparative and evolutionary studies applying to multigene families. In this context, we are developing the Receptor Tyrosine Kinase database (RTKdb), which is the only database on tyrosine kinase receptors presently available. In this database, protein sequences from eight model metazoan species are organized under the format previously used for the HOVERGEN, HOBACGEN and NUREBASE systems. RTKdb can be accessed through the PBIL (Pôle Bioinformatique Lyonnais) World Wide Web server at http://pbil.univ-lyon1.fr/RTKdb/, or through the FamFetch graphical user interface available at the same address.
INTRODUCTION
Receptor Tyrosine Kinases (RTK) form a protein superfamily, which is present in all metazoans, from sponge to humans. RTK are transmembrane proteins regulating key cellular processes during development and in adult-life, such as cell cycle, migration, metabolism, survival, proliferation and differentiation. They belong to the wider Tyrosine Kinase (TK) family, which is divided into two main groups: cytoplasmic Protein TK (PTK) and RTK. Function specificity of RTK among all other receptors comes from their tyrosine kinase activity that is induced following binding of a cognate ligand (1). RTK structure consists of an extracellular ligand-binding region, a transmembrane hydrophobic domain, and an intracellular region. Whereas extracellular region has variable length and sub-domain composition—thus defining RTK sub-families—the intracellular domain features the conserved kinase domain, sometimes split into two parts (TK1 and TK2) by a short amino acid sequence: the Kinase Insert (2) (Fig. 1A). Thus, RTKs can be represented as the arrangement of specific sub-domains, or modules—part of a sequence that have function unity or structure unity (3,4). Although secondary structure of the extracellular domain is conserved within a sub-family, another specificity level is represented by peculiar amino acids, which confer specificity to the ligand binding site. It is noteworthy that extracellular modules are varyingly shared between the various RTK sub-families, as well as between RTK and other membrane protein families. Similarly, the tyrosine kinase module is shared between RTK and PTK.
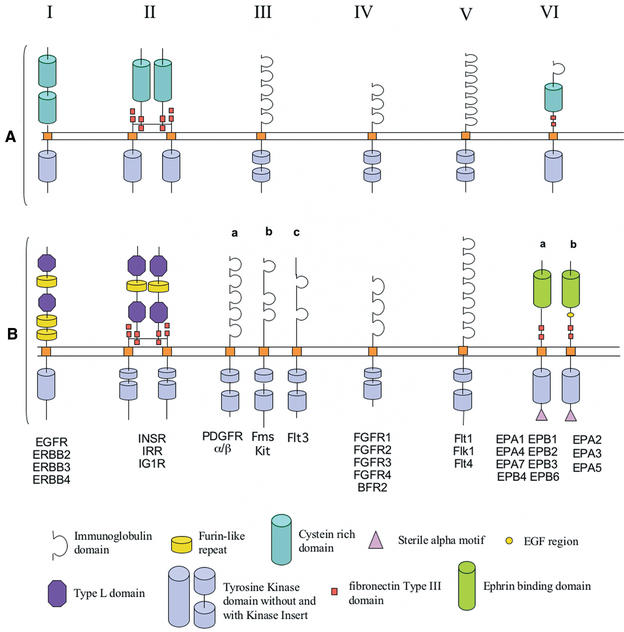
Figure 1.
(A) The first six RTK receptors classes among the 19 human RTK classes. We can observe that the extracellular region which is very different between all the RTKs, and, on the other hand, that the intracellular region is very conserved. (B) The new domain prediction of RTK. It shows some classification modifications and updated domain prediction from the latest domain databases SMART and Pfam.
RTKs exhibit modular structure and widespread expression. Moreover, their number and complexity increased during metazoan evolution, possibly in link with acquisition of new cellular function or regulatory processes. Thus, RTKs represent an excellent model for various evolutionary studies. In particular, it is not known whether RTK originated from a common ancestor and how RTK diversification occurred. RTK evolution may also imply ligand evolution. Within each RTK subfamily, cognate ligands share sequences similarities. This is especially the case for subclass III and subclass VI, suggesting coevolution of ligand-receptor pairs (5,6). For this reason, a future version of Receptor Tyrosine Kinase database (RTKdb) would include data on ligands. Finally, widespread distribution of RTK genes within the genome makes them a good model for investigating mechanisms implicated in genome evolution. However, despite a wealth of information on RTK, data are still dispersed among laboratories and no detailed and global phylogenetic analyzes have been performed so far on this class of sequences. To address these needs, we have developed RTKdb, a database dedicated to RTK sequences. This is the first database entirely devoted to that kind of molecules, it contains protein and DNA sequences, manually reviewed protein alignments and precomputed phylogenetic trees, as well as additional information such as module composition, or virtual expression patterns based on EST data. We have organized RTKdb data under the format presently used by different gene families databases already developed in GPs group: HOVERGEN (7), HOBACGEN (8) and NUREBASE (9). HOVERGEN and HOBACGEN are general gene families databases, gathering sequences from a wide range of organisms (vertebrates and prokaryotes, respectively), while NUREBASE is a more specialized system, devoted to nuclear receptor sequences.
DATABASE CONTENT
Release 0.3 (December 2001) of RTKdb contains 160 RTK protein entries taken from eight model metazoan species (Homo sapiens, Mus musculus, Rattus norvegicus, Gallus gallus, Danio rerio, Xenopus laevis, Drosophila melanogaster, Caenorhabditis elegans). The sequences in the database are not redundant. The database is built using sequences taken from SWISS-PROT (and soon, from its annex TrEMBL) (10). The reasons that led us to choose these collections are threefold: (i) the SWISS-PROT/TrEMBL set is exhaustive and almost non-redundant; (ii) SWISS-PROT annotations are of higher quality compared to other general database systems and (iii) almost all entries are cross-referenced with their corresponding nucleotide entries in GenBank/EMBL/DDBJ (11–13). RTKdb sequences are indexed through two ACNUC databases (14), one for the protein sequences in SWISS-PROT format, and one for the nucleotide sequences in EMBL format.
Original SWISS-PROT entries annotations are modified to include supplementary information (Fig. 2). Each entry has a sub-family accession number included in a new CC (comment) line within SWISS-PROT annotations. The sub-family accession number is manually assigned for the moment, but an automated annotation procedure is being developed. The format is RTKaabbbb, where aa corresponds to the species number (based on an alphanumerical numbering—10 numbers and 26 letters—to reach up to 1296 species), and bbbb to the sub-family number. For example, RTK010003 corresponds to a RTK belonging to the first species (H. sapiens) and the third sub-family class. When a protein sequence contains known ProDom domains (15), localization of these domains with starting and ending points and accession numbers are given in a corresponding FT (feature) line. ProDom was primarily used because it is presumably exhaustive. Indeed, ProDom is generated with an automated procedure applied to the whole SWISS-PROT database. Nevertheless, RTKdb was built with the idea of using diverse data sources for protein domain structures. For instance, we plan to integrate information provided by domain databases that are based on prediction with Hidden Markov Model (HMM) profiles such as SMART (16) or Pfam (17). Up to now, we have use these two other databases to update the extracellular domain prediction of human RTK. Indeed, for these proteins SMART and Pfam encompass a largest amount of information than other databases, and their use led us to change the classification of some families (Fig. 1B). For example, the extracellular domain of sub-family III was historically described as containing five immunoglobulin (Ig) repeats. We have discovered that the number of Ig repeats is in fact different between all the members of this subfamily, that led us to split it into three subfamilies (a, b, and c). This suggests an evolutionary history more complicated than the one previously established. Whenever possible, the predicted expression pattern is given for each receptor, following calculations using EST data (18).
Figure 2.
Annotations modifications of SWISS-PROT entries. The GENE_FAMILY field contains the RTKdb family number whereas the DOMAIN feature fields contain the locations of the different ProDom domains for the protein.
Multiple alignments and phylogenetic trees are computed for each sub-family. Protein sequences are aligned using CLUSTAL W (19), and then manually reviewed (especially for aligning very N-terminal protein sequences). All the default parameters are used except the ‘Toggle End Gap Separation’ option (in ‘Protein Gap Parameters’), which is turned on to optimise the alignment of sequences edges. Phylogenetic trees are also computed with CLUSTAL W, which implements the Neighbour Joining (NJ) method (20). NJ algorithm has been chosen for its speed, due to the unavoidable growth of the database in the near future. The distance used is the multiple substitutions correction, and trees are rooted by the mid-point method that minimizes the differences between branch lengths averages between the two sides of the root. Alignments and trees are stored into individual text files in CLUSTAL and NEWICK formats, respectively.
DATABASE ACCESS
Global organization of RTKdb uses the same structure as HOBACGEN or NUREBASE, with a client/server architecture. The server is made of three components: a World Wide Web service, a dedicated C program allowing to access the data, and the data (comprising the two ACNUC databases and text files containing sequences, alignments and trees). There are two ways to use RTKdb: through a World Wide Web server or through the FamFetch interface (8).
The aim of the RTKdb Web page is to centralize various data related to RTK that may be useful for the biologists' community and to add results produced by our own bioinformatics studies. The exploration starts from a single HTML document for each model organism. This page contains a clickable image representing a model of the structure of all the RTK available in this organism. From this page, it is possible to access either the list of sequences belonging to a given sub-family, the alignment, the tree or the expression data estimated through EST relative abundance. Sequences belonging to a family can be retrieved and downloaded easily through the on-line sequence database retrieval system WWW-Query (21). Alignments are displayed with the Jalview java applet, or can be downloaded as files picked up (in CLUSTAL format) by helper applications able to handle multiple alignments (for instance the program SeaView, see 22). Trees are displayed by the ATV java applet (23) or can also be downloaded by helper applications able to handle phylogenetic trees (like NJplot, see 24).
Access through FamFetch is devoted to a use for molecular evolution and genomic studies, whereas Web access is more oriented toward functional analysis studies. FamFetch is a Java 1.1 application, which allows interactivity and a wide portability on almost all platforms. The main window of the interface allows selection of one RTKdb sub-family (Fig. 3). It is also possible to write queries to define a subset of families matching specific criteria, such as species or keywords. Selection of a sub-family prompts a tree window with the corresponding phylogeny. In this tree, sequences are colored according to taxonomy. Four sets of twelve colour/taxon pairs are provided, and the user has the possibility to edit the color and/or the taxon used in each available pair. The tree display is active, with options of zooming, re-rooting, node swapping or subtree selection. Clicking on leaves allows selection of one or several receptors in a new window. From there, the user may view the DNA or protein entry, or the protein alignment. The alignment contains only the sequences selected by the user, and is not computed but reconstructed from the pre-existing whole family multiple alignment. Under version 2.0 of FamFetch, the user can also view the ProDom domains (if they are present) displayed as boxes, and check if they are correctly aligned (Fig. 3G). Functions allow the user to save lists of families, sequences entries, alignments or trees in text files.
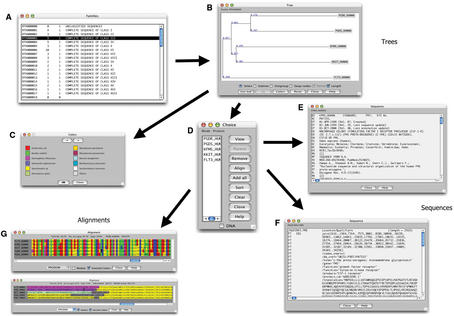
Figure 3.
Example of FamFetch session with RTKdb data. The ‘Families’ window allows one to perform queries and to select a given sub-family (A). Then, in the ‘Tree’ window, the phylogenetic tree associated with the selected sub-family is displayed (B). The ‘Colors’ dialog box shows the correspondence between colors and taxa (C). Cliking on a leaf on the ‘Tree’ window starts the display of the ‘Choice’ dialog box (D), from which it is possible to view: one of the selected sequence from SWISS-PROT (E) or EMBL (F) ACNUC databases. View of the alignment window with or without the ‘Module’ box checked (G).
DISCUSSION
Methods previously developed initially for collecting and organizing our fist set of RTK sequences will be fully automated, which will facilitate and accelerate RTKdb development. However, data obtained from the SWISS-PROT/TrEMBL databases should be analysed and authenticated by experts. Time lag between sequence publication and annotation require new tools for annotating sequences in order to face the increasing pace of genome sequencing. To extend RTKdb by accurate screening of new sequences, especially complete genomes, we will determine subclass-specific motifs within the intracellular region. New RTKs should be annotated with a specific method, yielding information suitable to bioinformatics as well as experimental use. Thus, additional informations are required, such as 3D structure, gene inactivation, mutation data and gene organization. Finally, in order to know more about genetic events leading to RTK diversification, information on genomic environment of RTK should be retrieved, accounting for gene duplication, recombination and translocation.
We are also working on an automated procedure that can strictly assign—on a weekly basis—new RTKs on their families from every common taxon and later, for every genome. With this significant amount of data, we will be able to start phylogenetic studies that could help us to better understand metazoan genome evolution. Moreover, due to our shared work with biologists we planned to add more specific biological information such as the nature of the ligands associated to each RTK or known mutations.
We also plan to use RTKdb to explore metazoan phylogeny. A preliminary study based on intracellular domains already confirmed the historical RTK classification, suggesting a long co-evolution between extra and intracellular regions (data not shown). It seems that recombination events leading to the present modular structure of extracellular domain are carrying the same phylogenetic information as the sequence mutations found in the intracellular domains. This also strengthens the concept of modular evolution of RTK. Some modules were probably acquired from other protein classes during evolution, resulting in the complex evolutionary story of RTK. Our analyses on the M. musculus and R. norvegicus data sets confirm the sub-family organization on close taxa (data not shown). But this is not exactly the same on remote taxa, particularly on nematode which possess sub-families close to human sub-families as well as distant sub-families (25). At last, extending phylogenetic studies to other metazoan species would help to describe RTK evolution more thoroughly.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge Claire Blachier and Frederic Tingaud for their helpful works. This work was supported by the ‘Centre National de la Recherche Scientifique’, and by a ‘Ligue Nationale Contre le Cancer’ grant. J.G. is recipient of a fellowship from the ‘Ministère de l'Education Nationale de la Recherche et de la Technologie’.
REFERENCES
- 1.van Geer P. and Hunter,T. (1994) Receptor Protein-Tyrosine Kinases and their signal transduction pathways. Annu. Rev. Cell Biol., 10, 251–337. [DOI] [PubMed] [Google Scholar]
- 2.Schlessinger J. (2000) Cell signaling by receptor tyrosine kinase. Cell, 103, 211–225. [DOI] [PubMed] [Google Scholar]
- 3.Patthy L. (1991) Modular exchange principles in proteins. Curr. Opin. Struct. Biol., 1, 351–361. [Google Scholar]
- 4.Patthy L. (1994) Introns and exons. Curr. Opin. Struct. Biol., 4, 383–392. [Google Scholar]
- 5.Drescher U. (2002) Eph family functions from an evolutionary perspective. Curr. Opin. Genet. Dev., 4, 397–402. [DOI] [PubMed] [Google Scholar]
- 6.Hannum C., Culpepper,J., Campbell,D., McClanahan,T., Zurawski,S., Bazan,J.F., Kastelein,R., Hudak,S., Wagner,J., Mattson,J. et al. (1994) Ligand for FLT3/FLK2 receptor tyrosine kinase regulates growth of haematopoietic stem cells and is encoded by variant RNAs. Nature, 368, 643–648. [DOI] [PubMed] [Google Scholar]
- 7.Duret L., Perrière,G. and Gouy,M. (1999) HOVERGEN: database and software for comparative analysis of homologous vertebrate genes. In Letovsky,S. (ed.), Bioinformatics Databases and Systems. Kluwer Academic Publishers, Boston, pp. 13–29.
- 8.Perrière G., Duret,L. and Gouy,M. (2000) HOBACGEN: database system for comparative genomics in bacteria. Genome Res., 10, 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duarte J., Perrière,G., Laudet,V. and Robinson,M. (2002) NUREBASE: database of nuclear hormone receptors. Nucleic Acids Res., 30, 364–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J., Rapp,B.A. and Wheeler,D.L. (2002) GenBank. Nucleic Acids Res., 30, 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoesser G., Baker,W., van den Broek,A., Camon,E., Garcia-Pastor,M., Kanz,C., Kulikova,T., Leinonen,R., Lin,Q., Lombard,V., Lopez,R., Redaschi,N., Stoehr,P., Tuli,M.A., Tzouvara,K. and Vaughan,R. (2002) The EMBL nucleotide sequence database. Nucleic Acids Res., 30, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tateno Y., Imanishi,T., Miyazaki,S., Fukami-Kobayashi,K., Saitou,N., Sugawara,H. and Gojobori,T. (2002) DNA Data Bank of Japan (DDBJ) for genome scale research in life science. Nucleic Acids Res., 30, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gouy M., Gautier,C., Attimonelli,M., Lanave,C. and di Paola,G. (1985) ACNUC—a portable retrieval system for nucleic acid sequence databases: logical and physical designs and usage. Comput. Appl. Biosci., 1, 167–172. [DOI] [PubMed] [Google Scholar]
- 15.Corpet F., Servant,F., Gouzy,J. and Kahn,D. (2000) ProDom and ProDom-CG: tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res., 28, 267–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letunic I., Goodstadt,L., Dickens,N.J., Doerks,T., Schultz,J., Mott,R., Ciccarelli,F., Copley,R.R., Ponting,C.P. and Bork,P. (2002) Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res., 30, 242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duret L. and Mouchiroud,D. (2000) Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol. Biol. Evol., 17, 68–74. [DOI] [PubMed] [Google Scholar]
- 19.Higgins D.G., Thompson,J.D. and Gibson,T.J. (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol., 266, 383–402. [DOI] [PubMed] [Google Scholar]
- 20.Saitou N. and Nei,M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol., 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 21.Perrière G. and Gouy,M. (1996) WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie, 78, 364–369. [DOI] [PubMed] [Google Scholar]
- 22.Galtier N., Gouy,M. and Gautier,C. (1996) SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci., 12, 543–548. [DOI] [PubMed] [Google Scholar]
- 23.Zmasek C.M. and Eddy,S.R. (2001) ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics, 17, 383–384. [DOI] [PubMed] [Google Scholar]
- 24.Kishino H., Miyata,T. and Hasegawa,M. (1990) Maximum likelihood inference of protein phylogeny and the origin of chloroplasts. J. Mol. Evol., 30, 151–160. [Google Scholar]
- 25.Popovici C., Roubin,R., Coulier,F., Pontarotti,P. and Birnbaum,D. (1999) The family of Caenorhabditis elegans tyrosine kinase receptors: similarities and differences with mammalian receptors. Genome Res., 9, 1026–1039. [DOI] [PubMed] [Google Scholar]