Abstract
The database PRODORIC aims to systematically organize information on prokaryotic gene expression, and to integrate this information into regulatory networks. The present version focuses on pathogenic bacteria such as Pseudomonas aeruginosa. PRODORIC links data on environmental stimuli with trans-acting transcription factors, cis-acting promoter elements and regulon definition. Interactive graphical representations of operon, gene and promoter structures including regulator-binding sites, transcriptional and translational start sites, supplemented with information on regulatory proteins are available at varying levels of detail. The data collection provided is based on exhaustive analyses of scientific literature and computational sequence prediction. Included within PRODORIC are tools to define and predict regulator binding sites. It is accessible at http://prodoric.tu-bs.de.
INTRODUCTION
The last decade has witnessed the successful completion of numerous bacterial genome-sequencing projects accompanied by their detailed annotation. Specialized databases on such widely studied model-organisms as Escherichia coli (1,2) and Bacillus subtilis (3,4), amongst others, reflect the added understanding of gene structure, expression and regulation. One central future target of bioinformatics is the integration of these data into regulatory networks. As yet, such integrated data and interpretative software are not widely available. Especially for the future understanding of the fine-tuned interaction between a bacterial pathogen and its host, it is necessary to store the existing knowledge in a structural database and to develop tools for modeling.
We, hereby describe a universal, genome-based database that describes and depicts prokaryotic gene regulation in great detail with a special emphasis on pathogenic bacteria. For this purpose, DNA binding sites of transcriptional regulators have been correlated with information on interacting proteins, promoter structures, operon and regulon organization by screening the original literature. The information on pathogenic bacteria is mapped onto their complete genome. We, additionally also provide a set of tools to predict DNA binding sites and to graphically depict genomic data.
Users are asked to cite this article when results were obtained using the database or tools therein.
STRUCTURE OF THE DATABASE
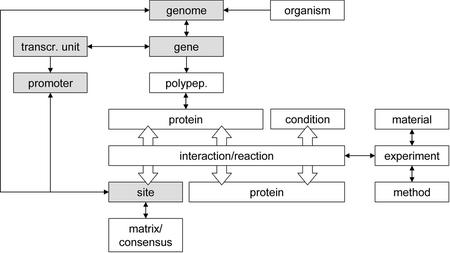
PRODORIC uses a relational database model. A modified TRANSFAC database structure (5) was gradually adapted to bacterial requirements (6). Figure 1 schematically depicts the relational structure of the main tables. The genomic sequence of the organisms included represents the structural backbone of the database. It is stored in a separate, numeric table. All DNA sequences described are linked to a fixed position within these genomes. Other genome-based tables such as Gene, Transcriptional Unit, Promoter and Binding Site similarly refer to a fixed and numeric positions in the appropriate genome. DNA sequence information, including ORF annotation and gene nomenclature, conforms to the standard style of the NCBI-server (http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/micr.html). The protein table, a major part of PRODORIC, contains functional and structural information on the proteome. The distinctive use of the terms ‘polypeptide’ and (functional) ‘protein’ permits the unambiguous discrimination of oligomeric protein complexes or heteromers from their constituent subunits. Proteins are classified according to the ‘cluster of orthologous groups’ (COG) classification (7). External databases are accessible through a direct link to the SWISS-PROT database (8).
Figure 1.
Schematic overview of the relational model core of the database structure, which describes protein--site- and protein--protein-interactions. Boxes in light grey refer to the corresponding genome sequence.
To distinguish protein–DNA, protein–protein and other interactions, we employ a central linking table. Numerous aspects of regulatory networks including external stimuli, promoter structures, regulatory factors and related metabolic pathways may thus be freely combined and used to analyse DNA binding sites, regulatory proteins, signal molecules or relevant metabolites. The future incorporation of regulatory circuits from pathogen-host interactions will further improve this powerful tool.
At present, we have restricted our annotational work to the structural definition of regulons. Information on established transcription factor—DNA binding site interactions was extended to include genes activated or repressed by these factors. Co-transcribed genes were designated a ‘transcriptional unit’. Experimental evidence was included through the Experiment table rated by confidence levels to reflect their reliability. In the description of protein-DNA-interactions, for example, data from DNAse-I-footprinting experiments were more highly rated than data from reporter gene fusion analyses. As indicated, regulatory DNA-motifs (sites) involved in transcription factor binding are linked to a fixed genomic location extending the annotated genome. To fully describe the promoter structure and transcriptional unit of each operon, this information was combined with features like transcriptional initiation sites and RNA polymerase binding sites and included in a separate promoter table. Where necessary, links to PubMed database are provided.
WORLD WIDE WEB INTERFACE AND ANALYSIS TOOLS
PRODORIC is available through a web interface (http://prodoric.tu-bs.de). Apart from the features outlined above, the website offers links to other molecular biology databases and bioinformatics tools. Forms and indexes facilitate user queries at our database. User requests are transformed into SQL queries by PHP scripts to generate dynamic web pages (9). The interface provides four forms reflecting the major biological questions: Genes/Operons, Proteins, Binding Sites and Matrices/Consensi. Each contains common search fields like ‘name’, ‘organism’, ‘description’ etc. Combined searches, use of wildcards and degenerated searches are also possible. Search results are tabulated. Links to external databases are provided where possible. Alternatively, an alphabetical list of regulons is available.
We have developed a new genome browser tool to provide an accessible overview of entire bacterial genomes. A subset of the genome may be displayed either as a schematic map in its genomic context or as a formatted, colour-coded sequence. In addition to the overall operon structure, the schematic map highlights sequence motifs and promoter information. Borders between coding and non-coding regions as well as transcription factor binding sites and other DNA sequences are clearly visible. Individual sequences may be exported by copy and paste. Navigation through the application is possible through a context-sensitive control panel.
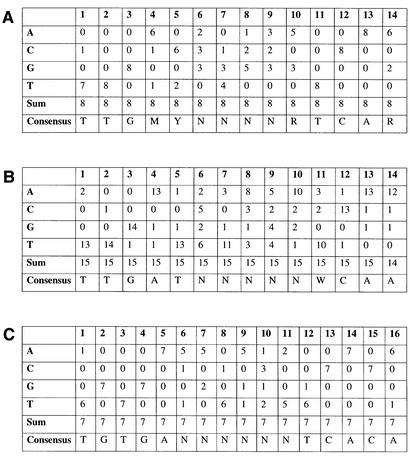
Regulator binding sites in promoter sequences may be predicted by scanning the sequence for putative binding motifs using a weight matrix representation. Alignment of binding sites allows the nucleotide distribution for every position to be determined (Fig. 2). The resulting distribution is used to locate additional DNA binding sites conforming to a user-defined threshold using our software tool Matrix Search based on published algorithms (10,11). Where possible, we have defined our own weight matrices for transcription factor binding sites. For several bacterial transcriptional regulators, only a single or a few individual DNA binding sites are known. Here a matrix representation is meaningless. The number of DNA sequences for matrix definition may be increased using orthologous binding sites of analogous regulators from other bacteria though slight differences in the binding consensi would introduce significant ambiguity. For example, an alignment of 15, 8 and 7 sequences, respectively from E. coli, Pseudomonas aeruginosa and B. subtilis indicate the binding site of the fumarate and nitrate reduction regulatory protein (Fnr, Fig. 2) to be clearly different in these organisms. The binding consensi in IUPAC 15-letter code are TTGMYNNNNRTCAR (E. coli), TTGATNNNNNWCAA (P. aeruginosa), and TGTGANNNNNNTCACA (B. subtilis). Our tool Consensus Search using simple IUPAC strings to define a binding motif consensus sequence is useful for those cases where only a few binding sequences are known. We provide a large library of weight matrices and binding consensi for bacterial transcription regulators. User-defined matrices and consensi may also be used.
Figure 1.
Weight matrix representations and resulting IUPAC consensi of the fumarate and nitrate reduction regulatory protein (Fnr) binding site and comparison between different species. (A) P. aeruginosa, (B) E. coli and (C) B. subtilis. The orthologous protein in P. aeruginosa is Anr (arginine fermentation and nitrate respiration regulator).
DATA CONTENT
We currently provide annotated genomes of five organisms though the data for the pathogenic bacteria P. aeruginosa, Listeria monocytogenes and Helicobacter pylori are most advanced while those for E. coli and B. subtilis were essentially collected to verify the tools provided. The data are accessible through web forms and the genome browser. PRODORIC is designed to be easily extensible allowing the facile incorporation of any microbial genome. The total number of entries is summarized in Table 1 (release September 2002), though this is expected to double by the time of publishing.
Table 1. Data content of PRODORIC (status as of September 2002).
| Species | Regulons | Sites |
|---|---|---|
| Pseudomonas aeruginosa | 25 | 89 |
| Listeria monocytogenes | 1 | 6 |
| Helicobacter pylori | 4 | 8 |
| Escherichia coli | 6 | 85 |
| Bacillus subtilis | 3 | 28 |
FUTURE PROSPECTS
Extensive screening of relevant literature is currently in progress to complete an up-to-date annotation of the P. aeruginosa genome. Other pathogenic bacteria are soon to follow. Additional transcription factor sites predicted by weight matrix searches will further extend the data. We have begun to include data provided by high throughput genomics and proteomics analyses to complement the published experimental data. We will shortly implement additional software tools and interconnected data tables containing information on signal transduction cascades and metabolic networks. The final step will be the description of pathogen-host interactions. We, therefore, intend to link our database to databases of eukaryotic regulators including TRANSFAC (5) and TRANSPATH (12).
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank the our colleagues Dirk Budke, Christoph Grunwald, Jörgen Haneke, Claudia Hundertmark and Johannes Klein for support in data annotation, literature scanning and programming and Dr Wolf-Dieter Schubert for critical reading of the manuscript. This work was funded by the German Bundesministerium für Bildung und Forschung (BMBF) for the bioinformatics competence center ‘Intergenomics’ (grant no. 031U110A/031U210A).
REFERENCES
- 1.Salgado H., Santos-Zavaleta,A., Gama-Castro,S., Millan-Zarate,D., Diaz-Peredo,E., Sanchez-Solano,F., Perez-Rueda,E., Bonavides-Martinez,C. and Collado-Vides,J. (2001) RegulonDB (version 3.2): transcriptional regulation and operon organization in Escherichia coli K-12. Nucleic Acids Res., 29, 72–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robison K., McGuire,A.M. and Church,G.M. (1998) A comprehensive library of DNA-binding site matrices for 55 proteins applied to the complete Escherichia coli K-12 Genome. J. Mol. Biol., 284, 241–254. [DOI] [PubMed] [Google Scholar]
- 3.Moszer I., Jones,L.M., Moreira,S., Fabry,C. and Danchin,A. (2002) Subtilist: the reference database for the Bacillus subtilis genome. Nucleic Acids Res., 30, 62–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishii T., Yoshida,K., Terai,G., Fujita,Y. and Nakai,K. (2001) DBTBS: a database of Bacillus subtilis promoters and transcription factors. Nucleic Acids Res., 29, 278–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wingender E., Chen,X., Fricke,E., Geffers,R., Hehl,R., Liebich,I., Krull,M., Matys,V., Michael,H., Ohnhäuser,R., Prüß,M., Schacherer,F., Thiele,S. and Urbach,S. (2001) The TRANSFAC system on gene expression regulation. Nucleic Acids Res., 29, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falb M. (2001) Etablierung einer Datenbank für prokaryontische Transcriptionsfaktoren (TRANSFACmicro). Diploma thesis, Technical University Braunschweig.
- 7.Tatusov R.L., Natale,D.A., Garkavtsev,I.V., Tatusova,T.A., Shankavaram,U.T., Rao,B.S., Kiryutin,B., Galperin,M.Y., Fedorova,N.D. and Koonin,E.V. (2001) The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res., 29, 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoll R.D. and Leierer,G.A. (2001) PHP 4+MySQL. Data Becker, Düsseldorf.
- 10.Schneider T.D., Stormo,G.D., Gold,L. and Ehrenfeucht,A. (1986) Information content of binding sites on nucleotide sequences. J. Mol. Biol., 188, 415–431. [DOI] [PubMed] [Google Scholar]
- 11.Quandt K., Frech,K., Karas,H., Wingender,E. and Werner,T. (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res., 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schacherer F., Choi,C., Goetze,U., Krull,M., Pistor,S. and Wingender,E. (2001) The TRANSPATH signal transduction database: a knowledge base on signal transduction networks. Bioinformatics, 17, 1053–1057. [DOI] [PubMed] [Google Scholar]