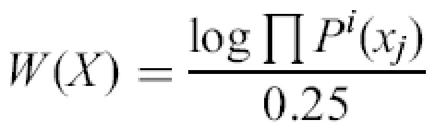Abstract
PlantProm DB, a plant promoter database, is an annotated, non-redundant collection of proximal promoter sequences for RNA polymerase II with experimentally determined transcription start site(s), TSS, from various plant species. The first release (2002.01) of PlantProm DB contains 305 entries including 71, 220 and 14 promoters from monocot, dicot and other plants, respectively. It provides DNA sequence of the promoter regions (−200 : +51) with TSS on the fixed position +201, taxonomic/promoter type classification of promoters and Nucleotide Frequency Matrices (NFM) for promoter elements: TATA-box, CCAAT-box and TSS-motif (Inr). Analysis of TSS-motifs revealed that their composition is different in dicots and monocots, as well as for TATA and TATA-less promoters. The database serves as learning set in developing plant promoter prediction programs. One such program (TSSP) based on discriminant analysis has been created by Softberry Inc. and the application of a support ftp: vector machine approach for promoter identification is under development. PlantProm DB is available at http://mendel.cs.rhul.ac.uk/ and http://www.softberry.com/.
INTRODUCTION
Draft nuclear genome sequences of Arabidopsis thaliana (1) and Oryza sativa (2,3), representing dicotyledonous and monocotyledonous higher plants, respectively, have been published. In addition, the putative gene contents of these genomes, predicted mostly by computer methods, are available (2,3,4; ftp://ftp.ncbi.nih.gov/genbank/genomes/A_thaliana; http://www.tigr.org/tdb/e2k1/ath1; http://mendel.cs.rhul.ac.uk/Arabidopsis). However, as both computer programs and experimental approaches for gene discovery have known limitations, we are still far from a fine picture of genome architecture. In particular, for all widely used gene prediction methods, one of the difficulties is accurate detection of the first (non-coding or partially coding) exon. The most accurate approach to solve this problem is to use information on full-length cDNAs. Unfortunately, no such information is available for most plant genes. Therefore, as well as being of special importance in understanding the regulation of gene expression, identification of plant promoters may serve as an essential element in gene annotation as well as in developing computational promoter prediction approaches. Currently, promoter identification is one of the most challenging problems in computational biology.
The term ‘promoter’ is used to designate a region in the genome sequence upstream of a gene transcription start site (TSS), although sequences downstream of TSS may also affect transcription initiation. Promoter elements select the transcription initiation point, transcription specificity and rate. Depending on the distance from the TSS, the terms of ‘proximal promoter’ (several hundreds nucleotides around the TSS) and ‘distal promoter’ (thousands and more nucleotides upstream of the TSS) are also used. Both proximal and distal promoters include sets of various elements participating in the complex process of cell-, issue-, organ-, developmental stage- and environmental factors-specific regulation of transcription. Most promoter elements regulating TSS selection are localized in the proximal promoter.
To date, there are a number of databases with information on cis-acting elements that control the transcription initiation by binding corresponding nuclear factors. These include TRANSFAC (5), TRRD (6), ooTFD (7), COMPEL (8), PlantCARE (9), PLACE (10) and RegSite (http://softberry.com). The last three databases are plant-oriented collections of transcription regulatory elements. The Eukaryotic Promoter Database (EPD) is only established collection of sequences of eukaryotic Pol II promoters (11). The latest release (#71) includes a total of 1402 entries, mainly of promoters from animals, with only about 200 from plant species.
In the course of development of a new computer method for predicting Pol II promoters of plant genes, we have collected Pol II promoter sequences from various plants. These data are incorporated on a new bioinformatics web server (http://www.mendel.cs.rhul.ac.uk) developed by the Department of Computer Science at Royal Holloway, University of London, in collaboration with Softberry Inc. (USA). It is designed to present information about plant genomes, genes and new approaches to their analysis. This article describes the criteria used for the promoter data collecting procedure, specific features of plant promoter sequences and Plant Promoter Database (PlantProm DB).
Description of PlantProm DB
Criteria for selecting promoter sequences
For collecting plant gene promoters the following rules were followed.
There is experimental evidence of the TSS position(s) of the gene, published in the literature. For genes with multiple TSSs the nearest to the CDS start position is taken, if no additional information on the predominance of one of them is available (positions of other TSSs are given in the name line of the sequence written in the FASTA format).
The length of known promoter sequence upstream of chosen TSS is 200 bp or more; all stored promoter sequences are the same length, 251 bp, where the position 201 corresponds to the TSS, i.e. collected sequences occupy the region (−200 : +51), with the TSS in the position +1, and, thus, present proximal promoters mentioned above.
An entry corresponds to the gene mapped on the genomic sequences.
Various alleles of a gene are presented in the database by a single entry.
Genes with more than one non-allelic copy in the genome as well as paralogous genes are taken as different entries.
Information content of the database
The annotated, non-redundant PlantProm DBL (release 2002.01) has 305 entries including 71, 220 and 14 promoters for RNA polymerase II from monocot, dicot and other plants, respectively. It provides the following information on plant promoters with experimentally known transcription start site(s):
DNA sequence of the promoter region (−200 : +51);
Nucleotide Frequency Matrices (NFM) for canonical promoter elements (TATA-box, CCAAT-box and TSS-motif or Initiator element, Inr);
Taxonomic and promoter type classification of promoters.
To compute nucleotide frequency matrices for various promoter elements, a pairwise comparison of a region [−50 : +1) of 305 plant promoters has been performed and one of the couple of promoters showing more than 90% homology has been excluded from the initial collection. As a result, 4 promoters were excluded and are denoted by ‘Excluded’ in the name line of these promoters sequences.
In simple implementation of Expectation Maximization (EM) algorithm (12), we considered the sequence of motif X=(x1,x2 ,… , xl), where l is the motif length. If Pi (xj) is the empiric frequency of the nucleotide xj in position i (computed on previous iteration), then the weight of this motif is computed as
Using the EM procedure for 10 iterations, the initial collection of 305 (301 unrelated) promoters was divided into the 2 classes: 175 (171 unrelated) TATA promoters and 130 TATA-less promoters. In calculations of TATA matrices, the allowed variation of a distance between the right boundary of the TATA-core box and the TSS was −18:−40 bp and only TATAWAWA-core was used for calculating the weight. As an initial TATA-box matrix, the TATA-matrix computed for 134 plant promoters from EPD (http://www.epd.isb-sib.ch/) was used. The computed TATA-matrix (Table 1) is in a good agreement with the TATA-matrix from EPD.
Table 1. Nucleotide frequencies matrix for TATA box from 171 unrelated plant promotersa.
| <2 | <1 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | >1 | >2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 0.28 | 0.16 | 0.03 | 0.95 | 0.00 | 1.00 | 0.62 | 0.97 | 0.38 | 0.73 | 0.13 | 0.30 |
| C | 0.27 | 0.63 | 0.01 | 0.00 | 0.04 | 0.00 | 0.00 | 0.00 | 0.01 | 0.08 | 0.42 | 0.42 |
| G | 0.17 | 0.05 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.02 | 0.00 | 0.10 | 0.28 | 0.16 |
| T | 0.28 | 0.16 | 0.96 | 0.05 | 0.96 | 0.00 | 0.38 | 0.01 | 0.61 | 0.09 | 0.18 | 0.11 |
| c | T | A | T | A | A/T | A | T/A | A |
aThe mean distance between TATA box and TSS is 26 bp.
For computation of the CCAAT-box matrix, we considered the possible distance between the right boundary of CCAAT-core and the TSS within −50:−100 bp. The CCAAT-core was used for weight calculation and, in accordance with the available data (13), CCAAT boxes were identified on both DNA strands. The CAAT matrix is presented in Table 2.
Table 2. Nucleotide frequencies matrix for CCAAT box from 131 unrelated plant promotersa.
| <4 | <3 | <2 | <1 | 1 | 2 | 3 | 4 | 5 | >1 | >2 | >3 | >4 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 0.31 | 0.34 | 0.27 | 0.30 | 0.31 | 0.00 | 1.00 | 1.00 | 0.00 | 0.28 | 0.32 | 0.29 | 0.40 |
| C | 0.19 | 0.17 | 0.16 | 0.18 | 0.34 | 1.00 | 0.00 | 0.00 | 0.00 | 0.20 | 0.20 | 0.25 | 0.17 |
| G | 0.20 | 0.20 | 0.27 | 0.21 | 0.15 | 0.00 | 0.00 | 0.00 | 0.00 | 0.20 | 0.18 | 0.15 | 0.15 |
| T | 0.30 | 0.29 | 0.30 | 0.31 | 0.20 | 0.00 | 0.00 | 0.00 | 1.00 | 0.32 | 0.30 | 0.31 | 0.28 |
| n | C | A | A | T |
aThe mean distance between CCAAT box and TSS is 75 bp.
The TSS-motif matrix of 5 bp in length has been computed, where the 3rd nucleotide was the annotated (anTSS). No strong consensus was revealed. When the EM approach was used to analyze all possible pentanucleotides with an assumed TSS (asTSS) location in the range (anTSS−2 : anTSS+2), it was observed that the composition of asTSS-motifs is different in dicot and monocot plants (Tables 3 and 4), as well as for TATA and TATA-less promoters (Tables 5 and 6). This finding seems to be a novel feature of plant promoters.
Table 3. Nucleotide frequencies matrix for a TSS-motif from 217 unrelated dicot plants' promotersa.
| −4 | −3 | −2 | −1 | +1 | +2 | +3 | +4 | |
|---|---|---|---|---|---|---|---|---|
| A | 0.341 | 0.249 | 0.286 | 0.005 | 0.604 | 0.475 | 0.226 | 0.272 |
| C | 0.184 | 0.286 | 0.041 | 0.507 | 0.332 | 0.028 | 0.359 | 0.240 |
| G | 0.101 | 0.124 | 0.041 | 0.161 | 0.065 | 0.101 | 0.129 | 0.198 |
| T | 0.373 | 0.341 | 0.631 | 0.327 | 0.000 | 0.396 | 0.286 | 0.290 |
| W | n | T/a | C/t | A/c | w |
aIn 75 cases, the high scoring TSS coincided with the annotated TSS.
Table 4. Nucleotide frequencies matrix for a TSS-motif from 70 unrelated monocot plants' promotersa.
| −4 | −3 | −2 | −1 | +1 | +2 | +3 | +4 | |
|---|---|---|---|---|---|---|---|---|
| A | 0.114 | 0.214 | 0.557 | 0.157 | 0.186 | 0.000 | 0.871 | 0.143 |
| C | 0.443 | 0.286 | 0.114 | 0.386 | 0.314 | 0.786 | 0.114 | 0.371 |
| G | 0.186 | 0.200 | 0.143 | 0.257 | 0.200 | 0.143 | 0.014 | 0.171 |
| T | 0.257 | 0.300 | 0.186 | 0.200 | 0.300 | 0.071 | 0.000 | 0.314 |
| a | N | n | C | A |
aIn 17 cases, the high scoring TSS coincided with the annotated TSS.
Table 5. Nucleotide frequencies matrix for a TSS-motif from 171 unrelated TATA promoters of plantsa.
| −4 | −3 | −2 | −1 | +1 | +2 | +3 | +4 | |
|---|---|---|---|---|---|---|---|---|
| A | 0.322 | 0.263 | 0.099 | 0.035 | 0.865 | 0.246 | 0.345 | 0.368 |
| C | 0.251 | 0.222 | 0.234 | 0.719 | 0.023 | 0.292 | 0.421 | 0.257 |
| G | 0.117 | 0.152 | 0.111 | 0.105 | 0.023 | 0.105 | 0.082 | 0.146 |
| T | 0.310 | 0.363 | 0.556 | 0.140 | 0.088 | 0.357 | 0.152 | 0.228 |
| T/c | C | A | n | M |
aIn 64 cases, the high scoring TSS coincided with the annotated TSS.
Table 6. Nucleotide frequencies matrix for a TSS-motif from 130 unrelated TATA-less promoters of plantsa.
| −4 | −3 | −2 | −1 | +1 | +2 | +3 | +4 | |
|---|---|---|---|---|---|---|---|---|
| A | 0.385 | 0.215 | 0.262 | 0.023 | 0.554 | 0.438 | 0.331 | 0.231 |
| C | 0.231 | 0.246 | 0.231 | 0.315 | 0.323 | 0.292 | 0.015 | 0.262 |
| G | 0.146 | 0.200 | 0.000 | 0.269 | 0.123 | 0.054 | 0.208 | 0.215 |
| T | 0.238 | 0.338 | 0.508 | 0.392 | 0.000 | 0.215 | 0.446 | 0.292 |
| T/a/c | Y | A/c | a/c/t | t/a/g |
aIn 46 cases, the high scoring TSS coincided with the annotated TSS.
PlantProm DB, release 2002.01, is available at the web sites http://mendel.cs.rhul.ac.uk and http://www.softberry.com. The database will be regularly updated by collection and analysis of new experimental data on plant promoters as it becomes available in the literature. PlantProm DB serves as a learning set in developing plant promoter prediction programs. One such program (TSSP), based on discriminant analysis of sequence features and plant regulatory motifs (RegSiteDB), has been developed by Softberry Inc. (http://www.softberry.com/berry.phtml?topic=promoter). The application of a support vector machine approach for promoter identification is under development.
Acknowledgments
ACKNOWLEDGEMENTS
PlantProm DB is funded by grant 111/BIO14428 ‘Pattern Recognition Techniques for Gene Identification in Plant Genomic Sequences’, from the UK Biotechnology and Biological Sciences Research Council (BBSRC) and is designed and maintained at Royal Holloway, University of London in collaboration with Softberry Inc. (USA).
REFERENCES
- 1.The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 2.Yu J., Hu,S., Wang,J., Wong,G.K., Li,S., Liu,B., Deng,Y., Dai,L., Zhou,Y., Zhang,X., Cao,M. et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science, 296, 79–92. [DOI] [PubMed] [Google Scholar]
- 3.Goff S.A., Ricke,D., Lan,T.-H., Presting,G., Wang,R., Dunn,M., Glaze-brook,J., Sessions,A., Oeller,P., Varma,H., Hadley,D. et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science, 296, 92–100. [DOI] [PubMed] [Google Scholar]
- 4.Schoof H., Zaccaria,P., Gundlach,H., Lemcke,K., Rudd,S., Kolesov,G., Arnold,R., Mewes,H.W. and Mayer,K.F. (2002) MIPS Arabidopsis thaliana Database (MAtDB): an integrated biological knowledge resource based on the first complete plant genome. Nucleic Acids Res., 30, 91–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wingender E., Chen,X., Fricke,E., Geffers,R., Hehl,R., Liebich,I., Krull,M., Matys,V., Michael,H., Ohnhäuser,R., Prüß,M., Schacherer,F., Thiele,S. and Urbach,S. (2001) The TRANSFAC system on gene expression regulation. Nucleic Acids Res., 29, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolchanov N.A., Ignatieva,E.V., Ananko,E.A., Pdkolodnaya,O.A., Stepanenko,I.L., Merkulova,T.I., Pozdyakov,M.A., Podkolodnny,N.L., Naumochkin,A.N. and Romashchenko,A.G. (2002) Transcription regulatory Regions Database (TRRD): its status in 2002. Nucleic Acids Res., 30, 312–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghosh D. (2000) Object-oriented Transcription Factors Database (ooTFD). Nucleic Acids Res., 28, 308–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kel-Margoulis O.V., Kel,A.E., Reuter,I., Deineko,I.V. and Wingender,E. (2002) TRANSCompel: a database on composite regulatory elements in eukaryotic genes. Nucleic Acids Res., 30, 332–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lescot M., Déhais,P., Thijs,G., Marchal,K., Moreau,Y., Van de Peer,Y., Rouzé,P. and Rombauts,P. (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res., 30, 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higo K., Ugawa,Y., Iwamoto,M. and Korenaga,T. (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res., 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Praz V., Périer,R., Bonnard,C. and Bucher,P. (2002) The eukaryotic promoter database, EPD: new entry types and linkes to gene expression data. Nucleic Acids Res., 30, 322–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardon L. and Stormo,G. (1992) Expectation maximization algorithm for identifying protein-binding sites with variable lengths from unaligned DNA fragments. J. Mol. Biol., 5, 159–170. [DOI] [PubMed] [Google Scholar]
- 13.Mantovani R. (1998) A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res., 26, 1135–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]