Abstract
The Sleeping Beauty (SB) transposon system provides the first random insertional mutagen available for germline genetic screens in mice. In preparation for a large scale project to create, map and manage up to 5000 SB insertions, we have developed the Mouse Transposon Insertion Database (MTID; http://mouse.ccgb.umn.edu/transposon/). Each insertion's genomic position, as well as the distance between the insertion and the nearest annotated gene, are determined by a sequence analysis pipeline. Users can search the database using a specified nucleotide or genetic map position to identify the nearest insertion. Mouse reports describe insertions carried, strain, genotype and dates of birth and death. Insertion reports describes chromosome, nucleotide and genetic map positions, as well as nearest gene data from Ensembl, NCBI and Celera. The flanking sequence used to map the insertion is also provided. Researchers will be able to identify insertions of interest and request mice or frozen sperm that carry the insertion.
INTRODUCTION
Insertional mutagenesis has proven an effective technique for the identification and initial functional characterization of genes, and is particularly well suited for organisms with a sequenced genome. In the fruit fly Drosophila melanogaster, endogenous transposons known as P-elements have long been used for insertional mutagenesis (1). With the sequencing of the Drosophila genome, the approach has proven particularly powerful. For example, the Berkeley Drosophila Genome Project (BDGP) gene disruption program has created insertions in over 1000 genes required for adult fly viability (2). These and other P-element insertions have been mapped onto the genome sequence and integrated into Flybase, allowing researchers to easily identify P-element insertions in or near genes of interest (3). The creation of a similar resource in mouse would aid mammalian gene identification and functional analysis of mammalian-specific, biomedically relevant traits. The release of the draft mouse genome by the Mouse Genome Sequencing Consortium (4) and the Sleeping Beauty (SB) transposon (5) now make this possible.
The SB transposon system was originally engineered from dormant Salmonid Tc1/mariner transposable elements (6). Although endogenous to fish, SB is active in all vertebrate cells tested in tissue culture and in mouse primary hepatocytes (7,8). More importantly, SB elements are active in the mouse male germline at a rate that makes it suitable for large-scale mutagenesis projects (5,9,10). A recent study has further indicated the utility of the SB system for large-scale mutagenesis. New germline insertions were obtained by breeding wild-type females to male mice that harbor both an SB transposase transgene and a multicopy transposon vector transgene. Offspring of such crosses inherit two new transposon insertions on average (5). A modified linker-mediated PCR method was used to clone and sequence a large number of the new transposon insertions in these offspring (C.M. Carlson, A.J. Dupuy, S. Fritz, C.F. Fletcher, K. Roberg-Perez, and D.A. Largaespada, manuscript submitted). Although half of the new insertions were located on chromosome 9, where the original multicopy transposon vector transgene is located, the remaining half were found scattered throughout the genome essentially at random. In addition, several of the insertions were located in or near known and predicted genes. Given these initial results, we have initiated a large-scale mutagenesis in the mouse using the SB system.
So that other researchers can take advantage of the SB insertions once they are mapped, we have developed the Mouse Transposon Insertion Database (MTID). MTID provides a web-based front end, which allows researchers to identify the insertion nearest any genomic position of interest. Researchers who identify a mouse with a desired insertion can request a mouse or frozen sperm and then investigate the functional role of the disrupted loci in mouse disease, physiology and development. We believe that the MTID will prove to be a useful resource for researchers studying mice and other mammalian systems.
MTID CONTENT
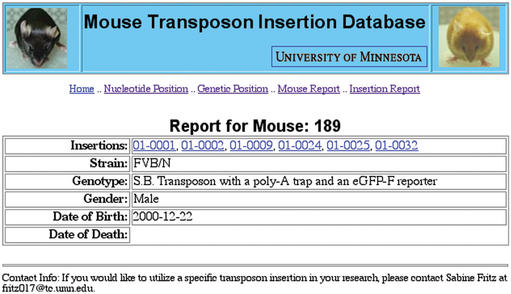
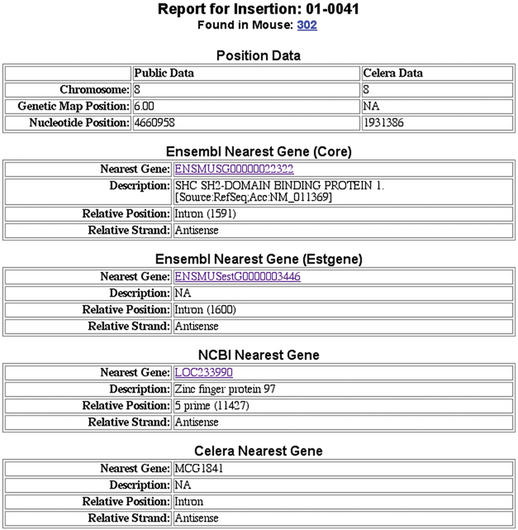
At the time of submission, the MTID comprises 44 SB-mediated transposon insertions from 26 mice. Data related to each mouse or insertion entity is provided to the user via report pages. These report pages are generated dynamically, providing the user with the most recent information. Data fields on the Mouse Reports include information on associated insertions, strain, genotype, gender, date of birth, and date of death (Fig. 1). On the Insertion Reports, the mouse or mice that carry the insertion are listed. Mapping information is provided for both public and Celera versions of the genome, including chromosome, genetic map position and nucleotide position (Fig. 2). In addition, the identity of the gene nearest to the given transposon insertion is provided for Ensembl, NCBI and Celera genome annotations. Nearest gene information fields include: nearest gene identifier, description (for NCBI genes this may include domain data), and relative position and distance to the transposon insertion. For each insertion, a comment is provided regarding the uniqueness of the BLAST hit used to assign the nucleotide position of the insertion (see Database Implementation below). There is also an associated Sequence Report that presents the flanking sequence used to determine the insertion's position and the border of the transposon from which the sequence was generated.
Figure 1.
A Mouse Report from MTID. Insertion identifiers are linked to the corresponding insertion report. The Genotype field describes the components of the Sleeping Beauty system carried by the given mouse.
Figure 2.
Detail of an MTID Insertion Report. Genomic position data are derived for both the public and Celera versions of the genome. Nearest gene data are determined for annotation information from Ensembl, NCBI and Celera. Predicted genes from Ensembl are currently divided into two classes: those confirmed based on similarity to EST sequences (Estgene) and those confirmed based on similarity to other sequences (Core). The distance between the insertion and the nearest gene is provided in parenthesis in the Relative Position field. When an insertion is identified within a gene this number represents the distance between the insertion and the nearest terminus of the gene.
MTID NAVIGATION
The MTID homepage (http://mouse.ccgb.umn.edu/transposon/) provides easy access to any part of the database. Links are provided under the MTID banner to search pages (by nucleotide or genetic map position) and to report retrieval pages (using mouse or insertion identifier). The home page also provides links to static pages that provide basic information including project background, a database introduction, relevant publications, methods used by the project, and contact information.
The search and retrieval pages allow users to access report pages of interest. Each is divided into a form that allows for data input and a brief description of how to use the form. The search pages require entry of a chromosome and either a nucleotide or genetic map position. Upon submission, an intermediate page is generated which reports the identity of the nearest insertion, the position of the insertion, and the distance between the user's position and the insertion. The nearest insertion identifier is linked to the corresponding Insertion Report. The retrieval pages are designed for users who have previously identified a mouse or insertion and wish to see an up-to-date report.
The Mouse and Insertion Reports are linked to each other as well as to additional data. On the Mouse Report, each insertion identifier acts as a link to the corresponding Insertion Report page (Fig. 1). Similarly, on the Insertion Report there are links to the associated mice (Fig. 2). Links are also available to the nearest Ensembl gene as well as to a view of the genomic region on the Ensembl site. A link to the BLAST report used to map the position is available, allowing researchers to examine the primary data used to map the insertion. In addition, a link to the corresponding sequence report is provided at the bottom of each Insertion Report.
MTID IMPLEMENTATION
The data model of MTID comprises 12 tables with mice and insertions treated as central objects. Currently, MySql 3.23.32 running on a 4-processor Sun Enterprise 450 is used for database management. All user access to the system is currently through the web interface. Pages are dynamically generated via Perl-CGI scripts, which integrate the results of queries on the database into various HTML templates. In addition to the public access pages, we have developed web editors that allow project members to enter new data not generated automatically. These tools have been designed to facilitate their reuse for other work performed by project members, such as analysis of proviral insertions.
We have developed an automated sequence analysis pipeline to populate the data fields derived from the public genome assembly and annotation. Such a system is necessary given the draft state of the mouse genome sequence, the increasing sophistication of genomic annotation, and our goal to create and map 5000 SB insertions. The pipeline uses resources from Ensembl (11), NCBI (12), the Mouse Genome Database (13) and BioPerl 1.0. The following describes the workflow of the pipeline.
BLAST (14) and BioPerl 1.0 are used to identify the contig positions of genomic sequences similar to the insertion flanking sequence.
The best BLAST hit is examined to estimate how reliable the resulting mapping may be. If the fraction of identical residues of the first BLAST hit is less than 0.95, the comment ‘Percent identity of best BLAST hit is below 95%’ is assigned and displayed on the Insertion Report. If the second blast hit is not sufficiently distinct from the first (if the match length is greater than or equal to 90% of the first, and the fraction of identical residues is greater than or equal to 95% of the first) the comment ‘Best blast hits are very similar’ is assigned. If neither of these points are true the comment ‘Distinct best blast hit’ is assigned.
The chromosome and nucleotide position of each insertion is determined by using the open database connection at Ensembl to access contig to chromosome mapping data.
Ensembl is again queried to identify the nearest SSLP marker. The marker is in turn used to estimate the genetic map position by querying marker and genetic map position data derived from the Mouse Genome Database.
Data from Ensembl and NCBI are queried to identify the known or predicted gene nearest the insertion.
The results are then deposited into the MTID back end and are immediately available to the public.
The sequence analysis pipeline significantly decreases the time required for annotating insertions. In 30 min a researcher can hand annotate only one insertion using a single annotation resource (such as the Celera Discovery System). In the same time our pipeline can process more than 50 insertions, incorporating data from both Ensembl and NCBI. Automation of this process will become increasingly important as MTID grows.
FUTURE DEVELOPMENTS
During the next year, we plan to identify and release several hundred new insertions. Additional fields on the Mouse Report will report insertion related phenotypes. Depending on user needs a search by gene function may be added as the number of insertions grows. To improve the sequence analysis pipeline, SSAHA (Sequence Search and Alignment by Hashing Algorithm) (15) will replace BLAST. Another data field may be created to report expression domains of disrupted genes using gene-trap transposon vectors designed to express a reporter protein such as GFP or LacZ. Related text and images are also being considered.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Sabine Fritz for providing the mouse images used on the MTID banner and Peter Fleck of the UMCC Informatics Core for assistance in developing the MTID web page format. We thank Chris Dwan for critically reading this manuscript and the University of Minnesota, Center for Computational Genomics and Bioinformatics for hosting MTID.
REFERENCES
- 1.Cooley L., Kelley,R. and Spradling,A. (1988) Insertional mutagenesis of the Drosophila genome with single P elements. Science, 239, 1121–1128. [DOI] [PubMed] [Google Scholar]
- 2.Spradling A.C., Stern,D., Beaton,A., Rhem,E.J., Laverty,T., Mozden,N., Misra,S. and Rubin,G.M. (1999) The Berkeley Drosophila genome project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics, 153, 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The FlyBase Consortium (2002) The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res., 30, 106–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindblad-Toh K., Lander,E.S., McPherson,J.D., Waterston,R.H., Rodgers,J. and Birney,E. (2001) Progress in sequencing the mouse genome. Genesis., 31, 137–141. [DOI] [PubMed] [Google Scholar]
- 5.Dupuy A.J., Fritz,S. and Largaespada,D.A. (2001) Transposition and gene disruption in the male germline of the mouse. Genesis, 30, 82–88. [DOI] [PubMed] [Google Scholar]
- 6.Ivics Z., Hackett,P.B., Plasterk,R.H. and Izsvak,Z. (1997) Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell, 91, 501–510. [DOI] [PubMed] [Google Scholar]
- 7.Izsvak Z., Ivics,Z. and Plasterk,R.H. (2000) Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J. Mol. Biol., 302, 93–102. [DOI] [PubMed] [Google Scholar]
- 8.Yant S.R., Meuse,L., Chiu,W., Ivics,Z., Izsvak,Z. and Kay,M.A. (2000) Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nature Genet., 25, 35–41. [DOI] [PubMed] [Google Scholar]
- 9.Fischer S.E., Wienholds,E. and Plasterk,R.H. (2001) Regulated transposition of a fish transposon in the mouse germ line. Proc. Natl Acad. Sci. USA, 98, 6759–6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horiee K., Kuroiwa,A., Ikawa,M., Okabe,M., Kondoh,G. et al. (2001) Efficient chromosomal transposition of a Tc1/mariner-like transposonSleeping Beauty in mice. Proc. Natl Acad. Sci. USA, 98, 9191–9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hubbard T., Barker,D., Birney,E., Cameron,G., Chen,Y., Clark,L., Cox,T., Cuff,J., Curwen,V., Down,T., Durbin,R., Eyras,E., Gilbert,J., Hammond,M., Huminiecki,L., Kasprzyk,A., Lehvaslaiho,H., Lijnzaad,P., Melsopp,C., Mongin,E., Pettett,R., Pocock,M., Potter,S., Rust,A., Schmidt,E., Searle,S., Slater,G., Smith,J., Spooner,W., Stabenau,A., Stalker,J., Stupka,E., Ureta-Vidal,A., Vastrik,I. and Clamp,M. (2002) The Ensembl genome database project. Nucleic Acids Res., 30, 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wheeler D.L., Church,D.M., Lash,A.E., Leipe,D.D., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Tatusova,T.A., Wagner,L. and Rapp,B.A. (2002) Database resources of the National Center for Biotechnology Information: 2002 update. Nucleic Acids Res., 30, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blake J.A., Richardson,J.E., Bult,C.J., Kadin,J.A., Eppig,J.T. and The Mouse Genome Database Group (2002) The Mouse Genome Database (MGD): the model organism database for the laboratory mouse. Nucleic Acids Res., 30, 113–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 15.Ning Z., Cox,A.J. and Mullikin,J.C. (2001) SSAHA: a fast search method for large DNA databases. Genome Res., 11, 1725–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]