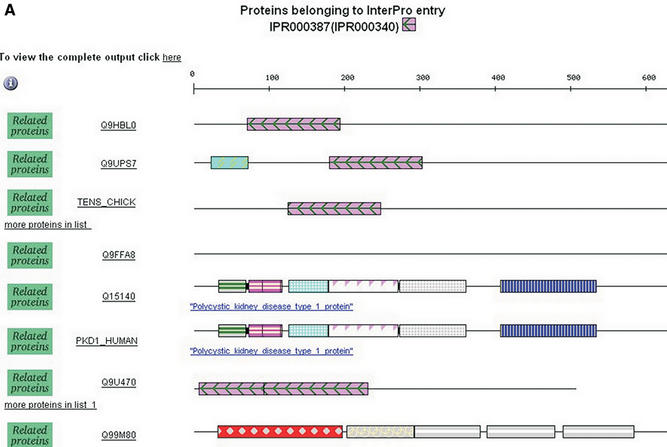
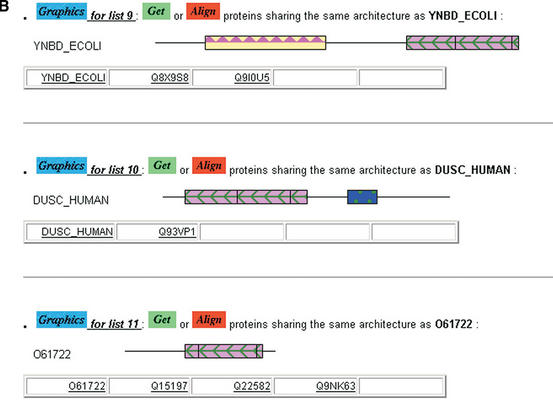
Nicola J Mulder
Nicola J Mulder
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
1,*,
Rolf Apweiler
Rolf Apweiler
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
1,
Teresa K Attwood
Teresa K Attwood
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
3,
Amos Bairoch
Amos Bairoch
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
4,
Daniel Barrell
Daniel Barrell
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
1,
Alex Bateman
Alex Bateman
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
2,
David Binns
David Binns
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
1,
Margaret Biswas
Margaret Biswas
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
5,
Paul Bradley
Paul Bradley
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
1,3,
Peer Bork
Peer Bork
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
6,
Phillip Bucher
Phillip Bucher
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
7,
Richard R Copley
Richard R Copley
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
8,
Emmanuel Courcelle
Emmanuel Courcelle
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
9,
Ujjwal Das
Ujjwal Das
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
1,
Richard Durbin
Richard Durbin
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
2,
Laurent Falquet
Laurent Falquet
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
7,
Wolfgang Fleischmann
Wolfgang Fleischmann
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
1,
Sam Griffiths-Jones
Sam Griffiths-Jones
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
2,
Daniel Haft
Daniel Haft
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
10,
Nicola Harte
Nicola Harte
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
1,
Nicolas Hulo
Nicolas Hulo
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
4,
Daniel Kahn
Daniel Kahn
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
9,
Alexander Kanapin
Alexander Kanapin
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
1,
Maria Krestyaninova
Maria Krestyaninova
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
1,
Rodrigo Lopez
Rodrigo Lopez
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
1,
Ivica Letunic
Ivica Letunic
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
6,
David Lonsdale
David Lonsdale
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
1,
Ville Silventoinen
Ville Silventoinen
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
1,
Sandra E Orchard
Sandra E Orchard
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
1,
Marco Pagni
Marco Pagni
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
7,
David Peyruc
David Peyruc
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
9,
Chris P Ponting
Chris P Ponting
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
11,
Jeremy D Selengut
Jeremy D Selengut
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
10,
Florence Servant
Florence Servant
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
1,
Christian J A Sigrist
Christian J A Sigrist
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
4,
Robert Vaughan
Robert Vaughan
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
1,
Evgueni M Zdobnov
Evgueni M Zdobnov
1EMBL Outstation—European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 3School of Biological Sciences and Department of Computer Science, The University of Manchester, Manchester, UK 4Swiss Institute for Bioinformatics, Geneva, Switzerland 5ViaLactia Biosciences, Newmarket Auckland, New Zealand 6Biocomputing Unit EMBL, Heidelberg, Germany 7Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland 8Wellcome Trust Centre for Human Genetics, Oxford, UK 9CNRS/INRA, Toulouse, France 10The Institute for Genomic Research, MD, USA 11MRC Functional Genetics Unit, Department of Human Anatomy and Genetics, University of Oxford, UK 12EMBL, Heidelberg, Germany
6,12