Abstract
The Mouse Genome Database (MGD) (http://www.informatics.jax.org) one component of a community database resource for the laboratory mouse, a key model organism for interpreting the human genome and for understanding human biology. MGD strives to provide an extensively integrated information resource with experimental details annotated from both literature and on-line genomic data sources. MGD curates and presents the consensus representation of genotype (sequence) to phenotype information including highly detailed information about genes and gene products. Primary foci of integration are through representations of relationships between genes, sequences and phenotypes. MGD collaborates with other bioinformatics groups to curate a definitive set of information about the laboratory mouse. Recent developments include a general implementation of database structures for controlled vocabularies and the integration of a phenotype classification system.
INTRODUCTION
The Mouse Genome Database (MGD) provides an integrated view of genetic and genomic information for the laboratory mouse (1). MGD contains information on mouse genes, genetic markers and genomic features as well as information on molecular segments (probes, primers, cDNA clones, BACs and YACs) mutant phenotypes, comparative mapping data, graphical displays of linkage, cytogenetic and physical maps, experimental mapping data, as well as strain distribution patterns for recombinant inbred strains (RIs) and cross haplotypes. MGD is updated daily (Table 1). Since it first became available on the WWW, MGD has continued to evolve, expanding its data coverage, improving data handling, and providing several new data manipulation and display tools.
Table 1. Snapshot of data content in MGD: September 23, 2002.
| MGD data statistics | September, 2002 |
|---|---|
| Number of references | 74 427 |
| Number of genes | 31 648 |
| Number of markers (including genes) | 51 261 |
| Number of genes with sequence data | 27 679 |
| Number of markers mapped | 41 258 |
| Number of mouse/human curator orthologies | 7488 |
| Number of genes with links to SWISS-PROT | 13 634 |
| Number of genes with GO annotations | 8574 |
| Number of genes with annotated alleles | 2760 |
| Number of annotated alleles | 7620 |
| Number of mouse nucleotide sequences curated and integrated in the MGI system (includes ESTs) | 540 898 |
MGD is one component of the Mouse Genome Informatics (MGI) database resource (http://www.informatics.jax.org) located at The Jackson Laboratory (http://www.jax.org). Other projects and resources that contribute to MGI include the Gene Expression Database (GXD) (2), the Mouse Genome Sequencing (MGS) (3) project and the Mouse Tumor Biology Database (MTB; http://www.informatics.jax.org/mtb) (4). The MGI consortium group participates actively in the development and implementation of the Gene Ontologies (GO) (www.geneontology.org) (5). MGI curators also collaborate extensively with SWISS-PROT (6) and with the LocusLink project at NCBI (7) to evaluate associations between genes and sequences for the mouse.
IMPROVEMENTS DURING 2002
Implementation of phenotype classifications
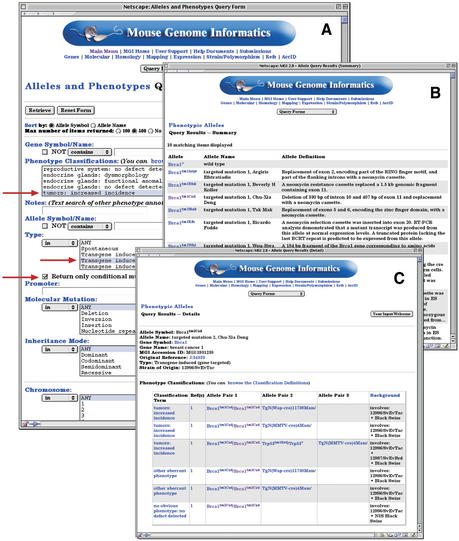
A broad, high-level set of phenotype terms have been developed and employed to classify phenotype data in MGD. This defined vocabulary of 105 terms can be used to search, group, compare and analyze phenotypes. These phenotype classification terms appear on the Alleles and Phenotypes Query Form (Fig. 1), and on the Genes and Marker Query Form. The complete list of terms and their accession IDs is also available by FTP. On each form, there is a link to the phenotype classification terms, complete with definitions and examples. Users of the MGI database can select one or more terms from the list to search for records associated with a particular phenotype, in combination with many other parameters on the forms. In addition, text-based searches for more specific phenotypic terms remain available.
Figure 1.
(A) The Allele and Phenotype Query Form permits queries based on Phenotype Classifications. In this example, the Phenotype Classification ‘tumors, increased incidence’ and Allele Type ‘Transgene induced (gene targeted)’ with only Conditional mutants returned. (B) The query presented in panel A returns a summary page of alleles defined by the criteria in A. In this case, 8 alleles met the criteria. The symbol, name and molecular description of these alleles are provided. Links are provided to the detail pages for each allele. (C) The top part of the detail page for the mutant phenotype of Brac1tm2Cxd details the Phenotype Classification terms for this particular mutation. Here, the details of the genotype that underlie the phenotype are represented. Each Phenotype Classification term is associated with one or more allele pairs and with the information known about the remaining genetic background. Thus, phenotypes are associated with genotypes, rather than simply with the allele.
A more comprehensive phenotype vocabulary continues to be developed by MGD staff and currently (September, 2002) contains over 1800 concepts. These terms are used to annotate mouse mutant phenotypes. Although these controlled terms are used to annotate mouse mutant phenotypes and can be viewed on allele detail pages, there currently is limited access to the full phenotype vocabulary as a query or analysis tool.
Improvements to the MGI : GO browser
The MGI GO Browser (http://www.informatics.jax.org/searches/GO_form.shtml) allows database users to access genes in MGI using functional annotation terms from the GO. This Browser was developed in conjunction with the GXD. A general database implementation within MGI for structured, controlled vocabularies enhances the search and recovery capabilities of this browser. The GO Browser can be accessed from gene detail or query pages as well as directly from the MGI menus. A GO Browser query returns a graph reflecting both parents and children of the query term and a link to all MGI associations with that term or any of the subterms.
Availability of MGI : GO files in various formats
MGI gene-to-GO annotations are updated daily. Various files for the MGI gene/markers with the GO associations are publicly available. These files are updated each time MGI submits a new gene association file to the GO web site (http://www.geneontology.org) and can be accessed on the MGI FTP server (ftp://www.informatics.jax.org/pub/informatics/reports/gene_association.mgi). A file of all the GO terms used by MGI in the annotation of genes and gene products is also available. MGI also provides a file to the GO database of MGI Gene : SWISS-PROT associations. This information is incorporated into the GO database and thus enables users to recover mouse sequence data as a result of a semantic search against the GO database (http://www.godatabase.org/cgi-bin/go.cgi).
OTHER INFORMATION
User input
MGD encourages user input into its gene and allele annotation efforts. On each gene detail and allele detail page, a clickable button (‘Your Input Welcome’) brings the user to a web-based form for submitting updates to the information being viewed.
Mouse gene nomenclature
The MGD gene annotation group assigns unique symbols and names to mouse genes under the guidelines set by the International Committee on Standardized Genetic Nomenclature for mouse (http://www.informatics.jax.org/mgihome/nomen/index.shtml#mnrg) (8). Scientists can reserve symbols prior to publication using the electronic nomenclature submission form (http://www.informatics.jax.org/mgihome/nomen/nomen_submit_form.shtml) or by contacting the MGD nomenclature coordinator by email (nomen@informatics.jax.org).
Electronic data submission
Any type of data that MGD maintains can be submitted as an electronic contribution, although mapping data, polymorphisms, and mammalian homologies are currently the most common. Each electronic submission receives a permanent database accession ID. All data sets are associated with either an electronic submission reference or a published paper. MGD reference pages provide links to associated data sets.
Community outreach and user support
MGD provides extensive user support through online documentation and easy email or phone access to User Support Staff.
User Support WWW access, http://www.informatics.jax.org/mgihome/support/support.shtml; Email: mgi-help@informatics.jax.org; Tel: +1 2072886445; Fax: +1 2072886132.
Other outreach
MGI-LIST (http://www.informatics.jax.org/mgihome/lists/lists.shtml), is a moderated and active email bulletin board supported by the MGI Users Support group. Other outreach includes Online Tutorials and answers to Frequently Asked Questions, available at: http://www.informatics.jax.org/userdocs/helpdocs_menu.shtml. Lee Silver's book, Mouse Genetics, is now available in an electronic version at http://www.informatics.jax.org/silver/. The online version has been enhanced by linking genes and references to MGI and MEDLINE.
IMPLEMENTATION
MGD is implemented in the Sybase relational database system, version 12.5. A large set of CGI scripts and Java Servlets mediate the user's interaction with the database. For computational users, direct SQL access can be requested through User Support. User-requested database reports and a number of widely used data files (generated daily) are available on the FTP site (ftp://ftp.informatics.jax.org).
CITING MGD
The following citation format is suggested when referring to datasets specific to the MGD component of MGI : Mouse Genome Database (MGD), Mouse Genome Informatics, The Jackson Laboratory, Bar Harbor, Maine (URL: http://www.informatics.jax.org). [Type in date (month, year) when you retrieved the data cited.]
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
MGD is supported by NIH/NHGRI grant HG00330. GO development and annotation efforts for MGI are supported by NIH/NHGRI grant HG02273.
REFERENCES
- 1.Blake J.A., Eppig,J.T., Richardson,J.E., Bult,C.J., Kadin,J.A. and Mouse Genome Database Group (2002) The Mouse Genome Database (MGD): the model organism database for the laboratory mouse. Nucleic Acids Res., 30, 113–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ringwald M., Eppig,J.T., Begley,D.A., Corradi,J.P., McCright,I.J., Hayamizu,T.F., Hill,D.P., Kadin,J.A. and Richardson,J.E. (2001) The mouse gene expression database. Nucleic Acids Res., 29, 98–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denny P. and Justice,M.J. (2000) Mouse as the measure of man? Trends Genet., 16, 283–287. [DOI] [PubMed] [Google Scholar]
- 4.Naf D., Krupke,D.M., Sundberg,J.P., Eppig,J.T. and Bult,C.J. (2002) The Mouse Tumor Biology Database: a public resource for cancer genetics and pathology of the mouse. Cancer Res., 62, 1235–1240. [PubMed] [Google Scholar]
- 5.The Gene Ontology Consortium (2001) Creating the Gene Ontology Resource: design and implementation. Genome Res., 11, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEML in 2000. Nucleic Acid Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acid Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maltais L., Blake,J.A., Chu,T., Lutz,C.M., Eppig,J.T. and Jackson,I. (2002) Rules and guidelines for mouse gene, allele, and mutation nomenclature: a condensed version. Genomics, 79, 471–474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.