Abstract
The University of Minnesota Biocatalysis/Biodegradation Database (UM-BBD, http://umbbd.ahc.umn.edu/) provides curated information on microbial catabolism and related biotransformations, primarily for environmental pollutants. Currently, it contains information on over 130 metabolic pathways, 800 reactions, 750 compounds and 500 enzymes. In the past two years, it has increased its breath to include more examples of microbial metabolism of metals and metalloids; and expanded the types of information it includes to contain microbial biotransformations of, and binding interactions with many chemical elements. It has also increased the ways in which this data can be accessed (mined). Structure-based searching was added, for exact matches, similarity, or substructures. Analysis of UM-BBD reactions has lead to a prototype, guided, pathway prediction system. Guided prediction means that the user is shown all possible biotransformations at each step and guides the process to its conclusion. Mining the UM-BBD's data provides a unique view into how the microbial world recycles organic functional groups. UM-BBD users are encouraged to comment on all aspects of the database, including the information it contains and the tools by which it can be mined. The database and prediction system develop under the direction of the scientific community.
INTRODUCTION
As the University of Minnesota Biocatalysis/Biodegradation Database (UM-BBD, URL=http://umbbd.ahc.umn.edu/) begins its eighth year of operation, the world is entering the post-genomic era. Moving its focus from genome sequencing, the molecular biology community is actively mining genomic, proteomic and metabolomic data, to better understand how organisms function at the molecular level. The UM-BBD has at last matured enough for such systematic analysis. The past two years have seen many changes to it besides an increase in database size. These include a change in its underlying database management system, additional ways to access the data and an increase in the types of information it contains. Additional changes will be made in the coming two years. These are intended to facilitate mining of UM-BBD data by the scientific community and are discussed in more detail below.
PRESENT STATUS
UM-BBD data content and methods, including data format, update and access, have been previously reported (1–3). The UM-BBD URL was changed to http://umbbd.ahc.umn.edu/ in early 2001. At the same time, all compound and reaction pages were reviewed and updated. In mid-2002, the database was transferred to MySQL (4). By the end of 2002, the UM-BBD will have grown to contain information on over 130 metabolic pathways, 800 reactions, 750 compounds and 500 enzymes.
Emphasis on elements
The US Department of Energy now supports the growth of the UM-BBD through its Microbial Genome Program (5). Under their sponsorship, we will be adding more examples of microbial biotransformations of metals, metalloids and metal chelators. A list of such pathways now in the UM-BBD is shown in Table 1.
Table 1. Metals, metalloids and metal chelators in the UM-BBD.
| Metals | Metalloids | Metal chelators |
|---|---|---|
| Mercury and organomercurials | Arsenic and organoarsenics | Nitrilotriacetate (NTA) |
| Selenium and organoseleniums | Silicon and organosilicanes | |
| Tin and organotins | ||
| Uranium | ||
| Chromium |
The most recent list of these is found at http://umbbd.ahc.umn.edu/metals.html.
The UM-BBD has traditionally focused on the metabolism of organic compounds, with some depiction of the organometals, organometalloids and toxic heavy metals listed in Table 1. However, microbes interact with and are increasingly discovered to transform, many of the chemical elements that constitute the earth. A recent UM-BBD project depicts the chemical elements using a periodic chart that organizes elements into a framework consistent with their use and transformation by microbes. The six major elements of a microbial cell are H, O, C, N, P and S. Microbial cells almost universally use the main group elements Na, K, Ca, Mg, Cl, Se, Zn and the transition metals V, Mo,W, Fe, Mn, Co, Ni and Cu. Increasingly, specialized microorganisms have been discovered to transform radionuclides, such as uranium and technetium; precious metals such as silver and gold; and metalloid compounds such as tellurium oxyanions. These transformations or binding interactions are now included in the UM-BBD, with a convenient access from periodic charts that organize and provide a common entry point to this information (URL=http://umbbd.ahc.umn.edu/periodic/). If one or more UM-BBD compounds contain the element, a link to a dynamic list of them is available on each element page. An example page for the element arsenic is shown in Figure 1.
Figure 1.
Excerpt from the UM-BBD webpage for the element arsenic. If users selects the ‘chemical properties’ link, they are transferred to the Web Elements (http://www.webelements.com/) page for the element; if they select the ‘UM-BBD compounds’ link, they are transferred to a dynamically-generated list of all UM-BBD compound pages that contain that element. The complete page is found at: http://umbbd.ahc.umn.edu/periodic/elements/as.html.
Paths to prediction
The ability to accurately predict biodegradation has enormous implications. Industries will continue to synthesize new materials faster than they, or regulatory agencies and academic researchers, can study their environmental fate. Many companies already invest significant resources to predict biodegradation of their new compounds to avoid commercialization of materials that are later found to be dangerous and subsequently need to be withdrawn from commerce.
To start on the path to prediction tools, structure-based searching was added to the UM-BBD in early 2002, based on the JChem toolkit from ChemAxon, Ltd (6). A user can either draw the compound to be searched for using a Java Applet or enter its SMILES string, then choose the type of structure search (similarity, substructure or exact) to do, and the similarity threshold to use. The structures of matching UM-BBD compounds are returned. Clicking on a returned structure leads to the corresponding UM-BBD compound page. From a compound page, one can go to UM-BBD reactions of, and biodegradation pathways for, the compound. This is a first step towards predicting a biodegradation pathway for compounds the UM-BBD does not contain, since the biodegradation pathways for UM-BBD compounds that are similar to the query compound can suggest ways the query might be degraded.
The next step in the prediction path is to analyze the reactions used by microorganisms to catabolically transform organic functions groups, identify the functional groups in an organic molecule representation, and transform each functional group, one at a time. This is guided prediction, since the user sees all possible transformations at each step and guides the process to its conclusion. Challenges include the implementation of transformation rules and incorporating functional group interactions.
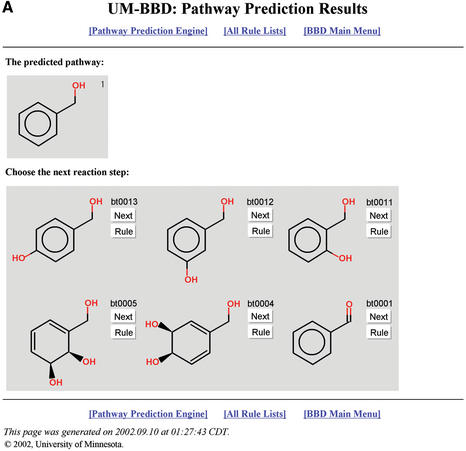
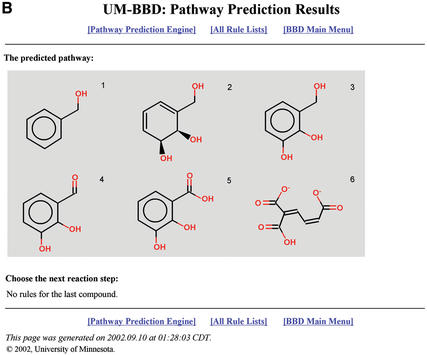
A prototype system with 40 rules (approximately one-third of those initially planned for the full system) was added to the UM-BBD in late 2002. A pathway prediction is shown in Figure 2; the first 10 rules are shown in Table 2. The JChem toolkit (6) and JOElib library (URL=http://joelib.sourceforge.net/) were used in construction of this system. With input from the scientific community, future work includes development and testing of additional transformation rules and determining how functional group interactions change rules.
Figure 2.
Excerpts from the UM-BBD Guided Pathway Prediction starting from the compound benzyl alcohol, see text. (A) First step, showing six possible biotransformations. The user proceeds by selecting ‘next’ to indicate the next compound to be transformed, or ‘rule’ to see the biotransformation (bt) rule for that transformation, including, where possible, a link to a UM-BBD reaction with bibliographic reference. (B) Last step in one predicted pathway branch. The compounds in the pathway are numbered 1–6, in pathway order. Additional rules are needed to handle the degradation of the last compound.
Table 2. Ten biotransformation formations rules for guided pathway prediction.
| bt0001: Primary alcohol→aldehyde |
| bt0002: Secondary alcohol→ketone |
| bt0003: Aldehyde→carboxylic acid |
| bt0004: Unsubstituted mono-aromatic→3,4 cis-dihydrodiol |
| bt0005: Unsubstituted mono-aromatic→1,2 cis-dihydrodiol |
| bt0006: 1,2 cis-dihydrodiol→1,2 dihydroxy catechol |
| bt0007: 3,4 cis-dihydrodiol→3,4 dihydroxy catechol |
| bt0008: 1,2 Dihydroxy→ring cleavage on one side of the two OH |
| bt0009: 1,2 Dihydroxy→ring cleavage between the two OH |
| bt0010: 1,2 Dihydroxy→ring cleavage on the other side of the two OH |
The most recent list of rules is found at: http://umbbd.ahc.umn.edu/predict/alllist.html.
CONCLUSIONS
Mining the UM-BBD's data provides a unique view into how the microbial world recycles organic functional groups. Similar to the way UM-BBD users are encouraged to recommend pathways to be added to the database, they are also encouraged to submit additional information on biotransformations of elements and to test the prediction program and comment on individual prediction rules. In this way, the database and prediction system grow under the direction of the scientific community.
Acknowledgments
ACKNOWLEDGEMENTS
This research was supported by the Office of Science (BER), U.S. Department of Energy, Grant No. DE-FG02-01ER63268. We thank Prasad Kotharu for assistance with the database and John Carlis, Tony Dodge, Steve Toeniskoetter and Jennifer Dommer for helpful discussions and other work on the biochemical periodic table.
REFERENCES
- 1.Ellis L.B.M., Hershberger,C.D. and Wackett,L.P. (1999) The University of Minnesota Biocatalysis/Biodegradation Database: specialized metabolism for functional genomics. Nucleic Acids Res., 27, 373–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellis L.B.M., Hershberger,C.D. and Wackett,L.P. (2000) The University of Minnesota Biocatalysis/Biodegradation Database: microorganisms, genomics and prediction. Nucleic Acids Res., 28, 377–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellis L.B.M., Hershberger,C.D., Bryan,E. and Wackett,L.P. (2001) The University of Minnesota Biocatalysis/Biodegradation Database: emphasizing enzymes. Nucleic Acids Res., 29, 340–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MySQL., Inc., 2510 Fairview Ave. East, Seattle, WA 98102.
- 5.US Department of Energy Microbial Genome Program, Oak Ridge National Laboratory (ORNL), 1060 Commerce Park MS 6480, Oak Ridge, TN 37830.
- 6.Csizmadia F. (2000) JChem: Java Applets and Modules Supporting Chemical Database Handling from Web Browsers. J. Chem. Inform. Comput. Sci., 40, 323–324. [DOI] [PubMed] [Google Scholar]