Abstract
Group II introns are self-splicing RNAs and retroelements found in bacteria and lower eukaryotic organelles. During the past several years, they have been uncovered in surprising numbers in bacteria due to the genome sequencing projects; however, most of the newly sequenced introns are not correctly identified. We have initiated an ongoing web site database for mobile group II introns in order to provide correct information on the introns, particularly in bacteria. Information in the web site includes: (1) introductory information on group II introns; (2) detailed information on subfamilies of intron RNA structures and intron-encoded proteins; (3) a listing of identified introns with correct boundaries, RNA secondary structures and other detailed information; and (4) phylogenetic and evolutionary information. The comparative data should facilitate study of the function, spread and evolution of group II introns. The database can be accessed at http://www.fp.ucalgary.ca/group2introns/.
INTRODUCTION
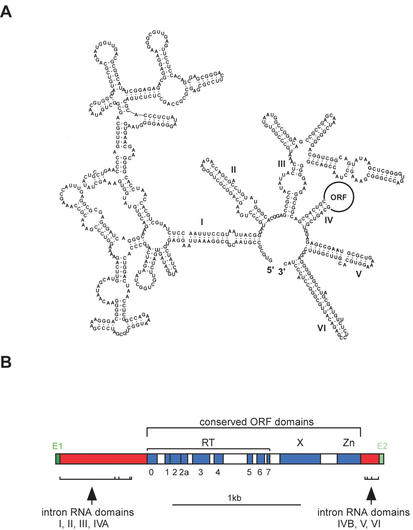
Group II introns are novel genetic elements that have properties of both catalytic RNAs and retroelements. The intron RNAs fold into a conserved structure with six domains (Fig. 1A), and are capable of catalyzing their own splicing reaction. Because of mechanistic and structural similarities, group II introns are believed to have been predecessors of spliceosomal introns (1,2).
Figure 1.
Structure of mobile group II introns. (A) RNA secondary structure. An example RNA secondary structure is shown for Novosphingobium aromaticivorans I1 (GenBank accession # AF079317), showing the six structural domains. When the intron encodes an ORF, it is looped out of domain IV. (B) Intron-encoded ORF structure. A typical ORF structure is shown. The largest domain (RT) contains the seven subdomains common to all reverse transcriptases (0–7). Additional domains are domain X, which contributes a splicing, or maturase, function, and the Zn domain, which has an endonuclease activity utilized in mobility. The Zn domain is absent from many bacterial intron ORFs. The six RNA structural domains surround the ORF and are flanked by exon sequences (E1 and E2).
Some group II introns also encode a reverse transcriptase (RT) ORF and are actively mobile (Fig. 1B). The intron-encoded ORF contains three domains: an RT domain that contributes reverse transcriptase activity, domain X with splicing, or maturase, activity and the Zn domain, which has an endonuclease activity. Activities of all three domains are utilized during intron mobility (3–5).
Mobility of group II introns occurs mainly by site-specific insertions (retrohoming), or to a lesser extent, by insertions into noncognate sites (retrotransposition). The mechanism of mobility, called target-primed reverse transcription, utilizes catalytic activities of both the intron RNA and RT. The intron reverse splices into one strand of the DNA target site, and then the Zn domain cleaves the other strand and the cleaved target DNA is used as a primer to reverse transcribe the intron (3–5). The mobility mechanism of group II introns is related to that of non-LTR elements (e.g. LINE elements), providing another link between group II introns and nuclear elements.
Group II introns were discovered and first characterized in organelles, where the majority of the introns are ORF-less and nonmobile. In bacteria, in contrast, group II introns almost always encode RT ORFs and are retroelements (6,7). The growing number of known group II introns in bacteria is stimulating new ideas about group II intron function and evolution. Currently, the number of known bacterial group II introns exceeds 40, and the number is certain to increase as more bacterial genomes are sequenced. However, most copies are not correctly identified, and there are also many fragments of group II introns in bacteria that confuse correct identifications.
Previously, we published work compiling and analyzing group II introns in bacteria and organelles, with analyses of secondary structures, intron-encoded ORFs, and phylogenetic relationships (6,8,9). We have now incorporated this information, as well as additional new data, into a web site that makes information on group II introns readily available to the public. We hope that this resource, which will be updated as information grows, will help correct many annotations of group II introns in the public databases, and will facilitate further study of group II intron function and evolution.
ORGANIZATION AND CONTENT OF THE DATABASE
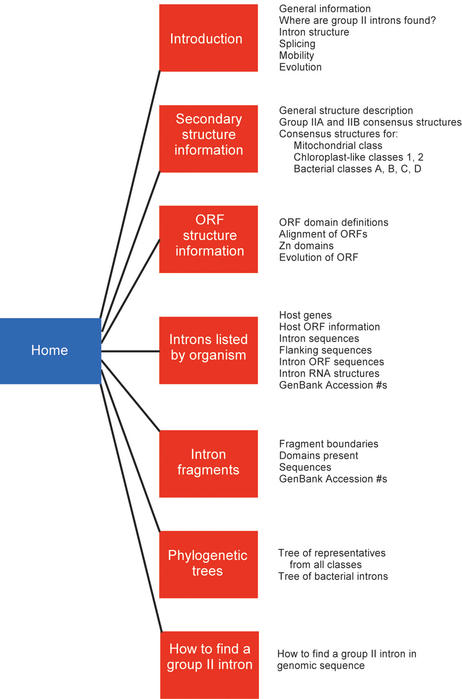
The organization of the database is diagrammed in Figure 2. An introductory section gives background information on group II introns, including their distribution in nature, their general RNA and ORF structures, their splicing and mobility mechanisms and models for their evolution. A separate section gives detailed information on the intron RNA structures, including the standard IIA and IIB structures and consensus structures for ORF-containing subclasses of intron RNAs. ORF structural information presented includes definitions of ORF domains, with the domain boundaries marked on an alignment of representative group II intron ORFs. The ORF alignment files can be downloaded as color PDF files.
Figure 2.
Organization of the web site.
The main data section consists of specific information listed for individual bacterial introns. A summary table lists introns with intron name abbreviations, host organisms and genes, ORF domains and sizes, classes of ORF and RNA structures and GenBank accession numbers. The links to intron names produces the specific sequence information for that intron. GenBank sequence with GenBank numbering is displayed, with red color-coding for the 5′ and 3′ ends of the intron and blue color-coding for the start and stop codons of the ORF. When the sequence is on the complementary strand of the annotated GenBank sequence, the complementary strand sequence is provided as well, but without numbering. A separate sequence entry displays the intron sequence with ∼400 bp of flanking sequence on each side of the intron. The ORF translation is also shown, as well as the specific intron RNA secondary structure.
A separate table gives equivalent information for bacterial intron fragments, which slightly outnumber full-length introns. The table lists the intron RNA and ORF domains present, and provides notes about the intron fragments (e.g. fragment boundaries, stem loops adjacent to the fragmentation site, flanking ORFs or elements, etc.) The numbered GenBank sequence is shown, with the same system of color-coding for the boundaries of the intron and ORF. The ORF translation contains highlighted amino acids showing the conserved motifs within the ORF fragment.
Phylogenetic trees based on RT ORFs divide the introns into seven classes: the mitochondrial class, chloroplast-like classes 1 and 2, and bacterial classes A, B, C and D (8). Each phylogenetic class also has a distinct RNA structure, although this is not the basis of the class definitions (9). An overall tree is shown which defines the classes using representative ORFs from mitochondrial, chloroplast and bacterial introns. This tree also provides information on how the ORF structure evolved, because it juxtaposes ORF domain features against the phylogenetic tree. The figure can be downloaded as a PDF file. A second tree is presented which includes nearly all bacterial intron ORFs, and omits the organellar introns.
An alignment of insertion sites for group II introns is provided, although there is little target site conservation even for related introns. Examples are also shown for introns that have multiple homing sites, as well as for the multiple insertion sites of bacterial class C introns, which insert directly after diverse transcriptional terminator sequences rather than into clearly defined homing site sequences. Finally, instructions are provided on how to find and identify group II introns in genomic sequence, which should be useful for persons new to group II introns who are trying to analyze genome sequences.
We hope that the information in this web site will provide a central pool of information on group II introns for those who are trying to identify and annotate new group II introns, as well as for group II intron researchers studying the function and evolution of group II introns. We believe there is much information to be found in the variety of group II introns. For example, some regions that are not conserved among all group II introns may be conserved within subclasses and contribute to class-specific functions.
FUTURE EXPANSION OF THE DATABASE
We plan to update the database periodically on an ongoing basis. There are sure to be many new introns reported in the next several years. In addition, we plan to add information on organellar ORF-containing introns, as the web site is currently focused on bacterial introns. Equivalent tables and information for organellar introns are in progress. We are considering adding information about ORF-less group II intron RNAs, as there is no public source for individual group II intron structures. Finally, we welcome corrections and additions to information in the web site, so that we can maintain accuracy and completeness.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by AHFMR (Alberta Heritage Foundation for Medical Research), CIHR (Canadian Institutes of Health Research) and NSERC (National Science and Engineering Research Council).
REFERENCES
- 1.Qin P.Z. and Pyle,A.M. (1998) The architectural organization and mechanistic function of group II intron structural elements. Curr. Opin. Struct. Biol., 8, 301–308. [DOI] [PubMed] [Google Scholar]
- 2.Michel F. and Ferat,J.-L. (1995) Structure and activities of group II introns. Annu. Rev. Biochem., 64, 435–461. [DOI] [PubMed] [Google Scholar]
- 3.Belfort M., Derbyshire,V., Parker,M.M., Cousineau,B. and Lambowitz,A.M. (2002) Mobile introns: pathways and proteins. In Craig,N.L., Craigie,R., Gellert,M. and Lambowitz,A.M. (eds), Mobile DNA II. ASM Press, Washington, DC, pp. 761–783.
- 4.Bonen L. and Vogel,J. (2001) The ins and outs of group II introns. Trends Genet., 17, 322–331. [DOI] [PubMed] [Google Scholar]
- 5.Lambowitz A.M., Caprara,M., Zimmerly,S. and Perlman,P.S. (1999) Group I and group II ribozymes as RNPs: clues to the past and guides to the future. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 451–485.
- 6.Dai L. and Zimmerly,S. (2002) Compilation and analysis of group II intron insertions in bacterial genomes: evidence for retroelement behavior. Nucleic Acids Res., 30, 1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez-Abarca F. and Toro,N. (2000) Group II introns in the bacterial world. Mol. Microbiol., 38, 917–926. [DOI] [PubMed] [Google Scholar]
- 8.Zimmerly S., Hausner,G. and Wu,X. (2001) Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res., 29, 1238–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toor N., Hausner,G. and Zimmerly,S. (2001) Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA, 7, 1142–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]