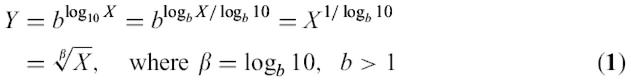Abstract
Recent enhancements and current research in the GeneCards (GC) (http://bioinfo.weizmann.ac.il/cards/) project are described, including the addition of gene expression profiles and integrated gene locations. Also highlighted are the contributions of specialized associated human gene-centric databases developed at the Weizmann Institute. These include the Unified Database (UDB) (http://bioinfo.weizmann.ac.il/udb) for human genome mapping, the human Chromosome 21 database at the Weizmann Insti-tute (CroW 21) (http://bioinfo.weizmann.ac.il/crow21), and the Human Olfactory Receptor Data Explora-torium (HORDE) (http://bioinfo.weizmann.ac.il/HORDE). The synergistic relationships amongst these efforts have positively impacted the quality, quantity and usefulness of the GeneCards gene compendium.
INTRODUCTION
Since 1997, GeneCardsTM (1–3) http://bioinfo.weizmann.ac.il/cards/, an automated, integrated database of human genes, genomic maps, proteins, and diseases, with software that retrieves, consolidates, searches, and displays human genome information, has enjoyed widespread popularity, and served as an impetus for similar projects in the field (4–7). Previous publications describe how the system has consistently added new features including sequence accessions, genomic locations, cDNA assemblies, orthologies, medical information, 3D protein structures and focused SNP summaries. Part of what continues to make GeneCards unique, is its synergistic relationship with a variety of research efforts at the Weizmann Institute. This paper describes the GeneCards (GC) features and algorithms that haven't previously been covered in the literature, as well as highlights the contributions of the companion, specialized databases United Database (UDB), Chromosome 21 database at the Weizmann Institute (CroW 21), and Human Olfactory Receptor Data Exploratorium (HORDE).
GeneCards
Enriched expression profiles
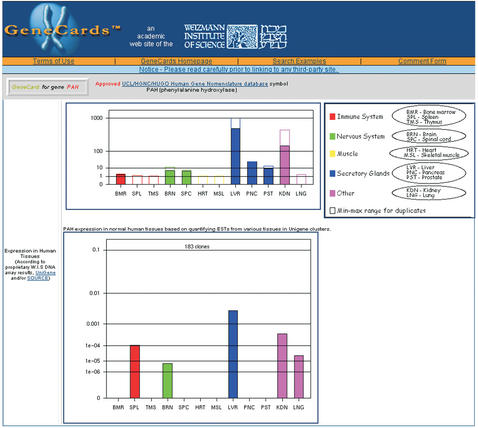
The construction of gene expression databases is a high priority for today's biological research community. Such databases, closely integrated with other types of genomic information, promise to both facilitate our understanding of many fundamental biological processes, and also accelerate drug discovery and customized diagnosis and treatment of diseases. In previous versions, the GC expression section provided a link to SOURCE (http://genome-www5.stanford.edu/cgi-bin/SMD/source/sourceSearch). GeneCards now additionally provides its own expression profiles that focus on normal human tissues. Two sets of tissue vectors are provided, one based on proprietary Weizmann Institute of Science experimental DNA array results, and the other based on in silico data mining and quantification of ESTs from a selected set of tissues in Unigene clusters (electronic Northern). GeneCards presentation of EST results differs from that of SOURCE in that GC applies heuristics to eliminate disease-related data, organizes results into color-coded groups (Fig. 1) and uses a root scale. Data on twelve tissues is presented; work is progressing on many others.
Figure 1.
Expression data in the GeneCard for PAH.
In the root scale used, the y-axis is computed as follows:
where X is the expression intensity value. This scale, designed solely for visualization purposes, enables viewing many orders of magnitude like on a logarithmic scale, but preserves the characteristic presentation of a linear scale in which the differences increase with the orders of magnitude. For b=2 (chosen for experimental tissue vectors), a 10-fold increase in expression doubles the corresponding Y value.
Experimental tissue vectors
Duplicate measurements were obtained for twelve normal human tissues (with the exception of pancreas where only one set of results is currently available) hybridized against Affymetrix GeneChip HG-U95A. Results were processed by the Affymetrix MAS4 program. Intensity values were normalized (currently without filtering or subtraction of background noise) and drawn on the described scale designed to ensure that a strong signal of a gene's transcripts in a particular tissue does not suppress the details of this gene's expression profile in other tissues. Currently, provided are tissue vectors for the 400 most popular GC genes (see the hot genes URL in the supplementary information).
Electronic Northern
For the same set of normal human tissues chosen for the experimental tissue vectors, NCBIs Unigene dataset (Hs.data) (8) was mined for information about the number of unique EST clones per gene per tissue. (The build number is noted on each card. The vectors are regenerated using the latest build each time GC is updated.) Clones are assigned to particular tissues by applying heuristics to the library information file (Hs.lib.info) to classify tissues into the chosen set, and to eliminate information not relevant to normal tissues (e.g. cancer data). Electronic expression results were calculated by dividing the number of EST clones per gene by the number of clones per tissue. Figure 1 also depicts this second type of expression profile, currently available for all GC genes that have associated Unigene records. Tissue vectors are presented with the same graphical rules as noted above in Equation 1. Here, the value b=√2 was chosen.
Tooltips are provided on mouseover to help explain the algorithms and scale.
Integrated gene locations
A new GC/UDB module, GeneLoc, integrates location data from different sources, eliminates redundancies, and provides a unique GC identifier based on genomic location. A problem inherent in mining and presenting information for every gene is that many genes are defined by high-throughput analysis of the whole genome, and lack functional data. Prediction programs may be applied to the genomic sequence to find potential genes, but results may vary depending on the particular program/parameters. Consequently, different sources may locate the same novel gene, but might not agree about its exact location and extent. Moreover, the gene is likely to be given a different name by each source. Sometimes, even known genes have a variety of names and positions.
GeneLoc uses information from Ensembl (9) and LocusLink (4), both based on the whole genome assembly of NCBI (8). Genes are first checked for identical HUGO symbols (10) and then for mutual links. If they share a HUGO symbol, they are assumed to be the same gene and assigned a single GC identifier. Similarly, if they share a LocusLink id, which implies that Ensembl and LocusLink have already associated these genes, they are merged and receive one GC identifier. When the chromosomal locations given by the two sources differ, both are associated with the merged gene. Next, GeneLoc tries to match by chromosomal position on both strands. An elaborate procedure is used in which gene locations from the two sources are compared, and an attempt is made to resolve cases of overlap of two or more genes (overlap clusters). When a gene from an overlap cluster shares no exons with any other gene from the cluster, it is given its own identifier. When more than two genes share exons, each is tagged with an attribute noting the exon sharing. The locations determined by this algorithm appear in GeneCards as absolute base coordinates.
UDB
Historically, GeneCards began as an offshoot of the Unified Database (UDB) (11) http://bioinfo.weizmann.ac.il/udb. UDB presented an integrated map for each human chromosome, based on data from various radiation hybrid, linkage and physical mapping resources, and further improved by anchorage of NCBIs (8) genomic contigs (containing finished and unfinished sequences), and repositioning of markers, genes and EST clusters in the sequenced regions. Due to its tabular output format and extensive set of genomic markers, UDB is especially useful for obtaining a bird's-eye view of genomic regions in linkage analysis and gene discovery projects.
Originally, the UDB map was based on Sequence Tagged Sites (STSs), genetic and radiation hybrid markers from different resources, combined using an interpolation algorithm into a single megabase scale map for each chromosome. Subsequently, EST clusters were anchored to the map according to their included markers. For clusters identified as known genes, the symbols were also included and linked to GC. Later versions applied a Sequence-Based Repositioning (SBR) algorithm, which incorporated genomic contig sequence data into the map, using it to generate more accurate relative positions.
A crucial step in building UDB was the establishment of a thesaurus for primary names and aliases of genomic markers. Marker names often differ among the sources; UDB combines them under a unique identifier. This thesaurus currently contains 154 120 markers. Further, UDB stores the primer sequences and product sizes of the STSs. Using the electronic PCR program (12) more than 100 000 STSs were placed on the sequence-based UDB map. About 8413 STSs were found to be associated with more than one location in the genomic sequences; these non-unique markers were rejected, unless they appeared only twice, with the two less than 1 Mb apart. A web-based form enables one to specify the region of interest as a cytogenetic band, a genomic interval or as genomic objects such as markers, genes or EST clusters. Both primary names and aliases may be entered as keywords. The selected map region is displayed in a customizable tabular format. Each row represents a location on the integrated map, with indicated UDB megabase coordinates. Each column shows objects of a different class—markers, genes or clusters. For markers, additional details appear, including DS-number, polymorphic nature, cDNA origin and specific map affiliation. For every marker, an internal link is provided to a MarkerCard which lists additional details retrieved from the sources, including original map data, full DNA sequence and flanking primers, synonymous names and heterozygote frequency for linkage markers.
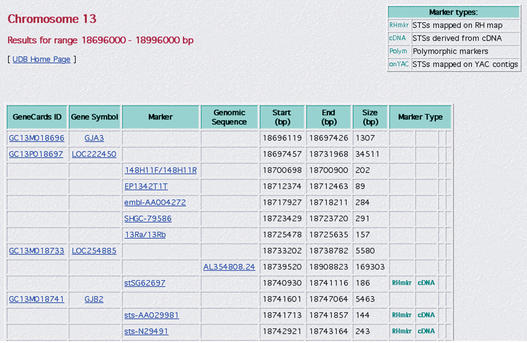
Figure 2 shows an extract of GeneCards GeneLoc results incorporated into UDB, integrating GeneCards numbers and gene positions with the markers and contigs that are already present in the database.
Figure 2.
GeneCards Integrated Gene Locations incorporated into UDB.
CROW 21
Chromosome 21 is the smallest human autosome, and was one of the first human chromosomes to be fully sequenced (13). Trisomy of chromosome 21 (phenotypically manifested as Down's syndrome) is the most frequent cause of mental retardation. Chromosome 21 contains important disease genes such as APP, RUNX1 (AML1), and SOD1. The latter was the first gene on chromosome 21 to be cloned and sequenced (14,15). The human CroW 21 data at the Weizmann Institute (CroW 21, http://bioinfo.weizmann.ac.il/crow21), combines the power of UDB, GeneCards and the GESTALT workbench (16), to provide a rich, easy-to-use view of chromosome 21. Chromosome 21 was retrieved from GenBank. 9.394 Mb of unsequenced region were added to the p-terminal, and 1.74 Mb of unsequenced region were added to the centromeric region. These estimates were based on data presented in NCBI's MapViewer. Annotations of gene locations were retrieved from GenBank, enabling accurate placement of genes on the map. STSs and polymorphic markers, retrieved from UDB, were placed on the sequence by e-PCR (12). Finally, the position of each GenBank genomic segment (AP001656–AP001761) was also noted on the chromosomal map.
Chromosome 21 was split into 250 kb sequence stretches that were input to GESTALT, a tool that integrates and automates the analysis of genomic sequences and produces annotated genomic map images (16). The identified genes are marked on the images, including the positions of their exons in both strands. Further, from the images, one can predict additional genes, alternative exons, promoters and so on. The CroW 21 site accesses the UDB and GC search engines, as well as the GESTALT visualizations. In turn, CroW 21 data is included in those databases. CroW 21s UDB backbone allows high-resolution exploration into the sequenced chromosome, providing information about the many genetic entities according to their chromosomal location.
HORDE
Olfactory receptors (ORs) constitute the largest multi-gene family in multi-cellular organisms. Their evolutionary proliferation has been driven by the need to provide recognition capacity for millions of potential odorants with arbitrary chemical configuration. The HORDE http://bioinfo.weizmann.ac.il/HORDE, is a database of human OR genes. It serves as a tool for studying phylogenetics, evolution and functionality of the OR gene and protein super-family. HORDE extracts its data directly from human genome sequence resources, using a semi-automatic data-mining procedure (17,18). A collection of 900 genes and pseudogenes were published (18). Another 116 genes, reported here for the first time, resulted from re-running the data-mining procedure and curating the results. These novel genes span all 17 families of the OR repertoire, where 16 of them opened new sub-families. Finally, to ensure completeness, we queried Celera's human genome assembly (19). Using 17 consensus sequences (one from each OR family) as input, we were able to detect 35 additional genes.
ORs tend to be disposed in clusters, a phenomenon accounted for by an elaborate process of gene and cluster duplication, as well as gene conversion events (20). To have an overview of these evolutionary processes, HORDE supplies information on genomic localization as well as cluster organization of the human OR repertoire. Genomic localization was done on the basis of the Aug 01 freeze of the UCSC genome assembly (21) using the BLAT server at UCSC (22). In parallel, we determined an accurate coordinate for each OR gene on the NCBI assembly (NT contigs) and UDB. The entire repertoire was analyzed to define OR clusters, using the criterion that two consecutive ORs that are more than 0.8 Mb apart belong to different clusters. The C@M nomenclature of clusters is used, where C is the chromosome, and M the megabase coordinate on it (17). In the spirit of GeneCards, HORDE summarizes comprehensive information about each gene into a single OR card, which includes HUGO symbol, gene family and subfamily, aliases, cytogenetic band, closest mouse OR gene, genomic sources, nucleic and protein sequences, and localization and cluster membership, with links to GC, UDB, UCSC, ORBD and NCBI. An example of a HORDE OR card is shown in the supplementary information. HORDE offers several tools for data-retrieval: (i) textual, e.g. to access a specific OR card using a HUGO symbol or alias; (ii) a BLAST (8) server—to search HORDE with a sequence, resulting in a report, linked to HORDE cards; (iii) group oriented—most suitable for studying the evolution and phylogenetics of this huge super-family. ORs that belong to the same family and/or subfamily, or are located on the same chromosome, can be queried. The results can be further analyzed on-line using CLUSTALW (23). Finally, the site is equipped with other useful tools, such as conceptual translation (24), and recognition of transmembrane domains and CDR residues.
IMPLEMENTATION
All of the software is written in Perl. Data retrieval is by extraction from files mirrored locally, directly from source databases, or via remote queries. UDB data is stored in a SYBASE relational database, accessed using Perl DBI. The GeneLoc module uses MySQL. GeneCards text data files are being migrated to use XML.
SUPPLEMENTARY INFORMATION
Hot GeneCards genes: http://bioinfo.weizmann.ac.il/cards/hotCards.html. A sample HORDE card: http://bioinfo.weizmann.ac.il/cgi-bin/HORDE/showgene.pl?key=symbol&value=OR1F1. A sample MarkerCard: http://bioinfo.weizmann.ac.il/cgi-bin/udb/object_details_mkr_sbr.pl?id=130973&disp=html.
Acknowledgments
ACKNOWLEDGEMENTS
The work described in this paper was supported by the Weizmann Institute of Sciences Crown Human Genome Center and by the Abraham and Judith Goldwasser foundation.
REFERENCES
- 1.Rebhan M., Chalifa-Caspi,V., Prilusky,J. and Lancet,D. (1997) GeneCards: integrating information about genes, proteins and diseases. Trends Genet., 13, 163. [DOI] [PubMed] [Google Scholar]
- 2.Rebhan M., Chalifa-Caspi,V., Prilusky,J. and Lamncet,D. (1998) GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics, 14, 656–664. [DOI] [PubMed] [Google Scholar]
- 3.Safran M., Solomon,I., Shmueli,O., Lapidot,M., Shen-Orr,S., Adato,A., Ben-Dor,U., Esterman,N., Rosen,N., Peter,I. et al. (2002) GeneCardsTM 2002: towards a complete, object-oriented, human gene compendium. Bioinformatics, 11, 1542–1543. [DOI] [PubMed] [Google Scholar]
- 4.Pruitt K.D., Katz,K.S., Sicotte,H. and Maglott,D.R. (2000) Introducing RefSeq and LocusLink: Curated human genome resources at the NCBI. Trends Genet., 16, 44–47. [DOI] [PubMed] [Google Scholar]
- 5.Eckman B.A., Kosky,A.S. and Laroco,L.A.,Jr (2001) Extending traditional query-based integration approaches for functional characterization of post-genomic data. Bioinformatics, 17, 587–601. [DOI] [PubMed] [Google Scholar]
- 6.Gilbert D.G. (2002) euGenes: a eukaryote genome information system. Nucleic Acids Res., 30, 145–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lenhard B., Hayes,W.S. and Wasserman,W.W. (2001) GeneLynx: A gene-centric portal to the human genome. Genome Res., 11, 2151–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler D.L., Church,D.M., Lash,A.E., Leipe,D.D., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Tatusova,T.A., Wagner,L. et al. (2002) Database resources of the National Center for Biotechnology Information: 2002 update. Nucleic Acids Res., 30, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hubbard T., Barker,D., Birney,E., Cameron,G., Chen,Y., Clark,L., Cox,T., Cuff,J., Curwen,V., Down,T., et al. (2002) The Ensembl genome database project. Nucleic Acids Res., 30, 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wain H.M., Lush,M., Ducluzeau,F. and Povey,S. (2002) Genew: the Human Gene Nomenclature Database. Nucleic Acids Res., 30, 169–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chalifa-Caspi V., Rebhan,M., Prilusky,J. and Lancet,D. (1997) The Unified DataBase (UDB): a novel genome integration concept. Genome Digest., 4, 15–16. [Google Scholar]
- 12.Schuler G.D. (1997) Sequence mapping by Electronic PCR. Genome Res., 7, 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hattori M., Fujiyama,A., Taylor,T.D., Wantanabe,H., Yada,T., Park,H.S., Toyoda,A., Ishii,K., Totoki,Y., Choi,D.K. et al. (2000) The DNA sequence of human chromosome 21. Nature, 405, 311–319. [DOI] [PubMed] [Google Scholar]
- 14.Lieman-Hurwitz J., Dafni,N., Lavie,V. and Groner,Y. (1982) Human cytoplasmic super-oxide dismutase cDNA clone: A probe for studying the molecular biology of Down's syndrome. Proc. Natl Acad. Sci. USA, 79, 2808–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levanon D., Lieman-Hurwitz,J., Dafni,N., Wigderson,M., Sherman,L., Bernstein,Y., Laver-Rudich,Z., Danciger,E., Stein,O. and Groner,Y. (1985) Architecture and anatomy of the chromosomal locus in human chromosome 21 encoding the Cu/Zn superoxide dismutase. EMBO J., 4, 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glusman G. and Lancet,D. (2000) GESTALT: A workbench for automatic integration and visualization of large-scale genomic sequence analyses. Bioinformatics, 16, 482–483. [DOI] [PubMed] [Google Scholar]
- 17.Glusman G., Bahar,A., Sharon,D., Pilpel,Y., White,J. and Lancet,D. (2000) The olfactory receptor gene superfamily: data mining, classification, and nomenclature. Mamm. Genome, 11, 1016–1023. [DOI] [PubMed] [Google Scholar]
- 18.Glusman G., Yanai,I., Rubin,I. and Lancet,D. (2001) The complete human olfactory subgenome. Genome Res., 11, 685–702. [DOI] [PubMed] [Google Scholar]
- 19.Celera Discovery System (http://www.celera.com).
- 20.Trask B.J., Massa,H., Brand-Arpon,V., Chan,K., Friedman,C., Nguyen,O.T., Eichler,E., Van den Engh,G., Rouquier,S., Shizuya,H. et al. (1988) Large multi-chromosomal duplications encompass many members of the olfactory receptor gene family in the human genome. Hum. Mol. Genet., 7, 2007–2020. [DOI] [PubMed] [Google Scholar]
- 21.Human Genome Project Working Draft at the UCSC (http://genome.ucsc.edu).
- 22.Kent W.J. (2002) BLAST—the BLAST-like alignment tool. Genome Res., 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins D.G., Thompson,J.D. and Gibson,T.J. (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol., 266, 383–402. [DOI] [PubMed] [Google Scholar]
- 24.Pearson W., Wood,T., Zhang,Z. and Miller,W. (1997) Comparison of DNA sequences with protein sequences. Genomics, 46, 24–36. [DOI] [PubMed] [Google Scholar]