Abstract
The SNP Consortium website (http://snp.cshl.org) has undergone many changes since its initial conception three years ago. The database back end has been changed from the venerable ACeDB to the more scalable MySQL engine. Users can access the data via gene or single nucleotide polymorphism (SNP) keyword searches and browse or dump SNP data to textfiles. A graphical genome browsing interface shows SNPs mapped onto the genome assembly in the context of externally available gene predictions and other features. SNP allele frequency and genotype data are available via FTP-download and on individual SNP report web pages. SNP linkage maps are available for download and for browsing in a comparative map viewer. All software components of the data coordinating center (DCC) website (http://snp.cshl.org) are open source.
INTRODUCTION
The SNP Consortium (TSC) was established in 1999 as a collaboration of several companies and institutions to produce a public resource of single nucleotide polymorphisms (SNPs) in the human genome. The initial goal was to discover 300 000 SNPs in two years, but the final results exceeded this, as 1.4 million SNPs had been released into the public domain at the end of 2001 (1).
The TSC Data Coordinating Center (DCC) website (http://snp.cshl.org), maintained by our group at Cold Spring Harbor Laboratory, was established to make TSC project data available to the research community and provide information about the project itself. Initially, the website consisted of FTP access to whole database dumps and other useful tab-delimited datafiles, as well as an HTTP-based graphical interface for browsing the data, with ACeDB/AcePerl (2) serving as the database back end. The web browsing interface was completely redesigned in 2001 to use a more scalable MySQL database back end, though the website appearance stayed much the same. A Java applet allows users to view individual traces from the reduced representation shotgun sequencing, used to call individual SNPs. A new chromosome browsing interface was introduced at the end of 2001. This interface showed SNPs in context of gene predictions from the Ensembl project (3) and NCBI RefSeq mRNAs (4) mapped by our group onto the human draft genome assembly available at that time (5).
Now that the SNP discovery phase of the TSC project is essentially complete, the emphasis has shifted to studying SNPs in populations. Accordingly, new features have been introduced to the TSC website, not only to make the new population data accessible, but also to improve existing data browsing and searching facilities.
THE TSC DATA REPOSITORY AND DATA RELEASES
The TSC data repository contains data submitted by participating labs to the DCC pertaining the SNP discovery process, such as flanking sequences, traces used in the discovery, lab submitter, alleles found and so forth. More recently, results from the ongoing TSC allele frequency and genotype project have been submitted to the DCC and added to the repository. See Table 1 for a summary of the publicly available TSC data. These SNPs have all been submitted to dbSNP by the DCC. In addition, data on SNP allele frequencies in several populations have been submitted to dbSNP directly by labs involved in the TSC allele frequency/genotype project (6).
Table 1. Key statistics for the TSC database.
| Total SNPs available | 1 793 201 |
| Total SNPs with reported allele frequencies | 95 861 |
| Total allele frequency records | 242 987 |
| Total SNPs genotyped | 35 574 |
| Total genotypes | 4 579 012 |
| Total individuals genotyped | 999 |
At the time of writing, several hundred thousand SNPs in the primary database are neither available publicly on our website nor via dbSNP, either because they have not been made available in the first place or they were made available and later withdrawn because they do not map to a unique position in the genome. Recognizing that these SNPs are still potentially valuable, the DCC released those SNPs in fall 2002.
During the main TSC SNP discovery phase, complete data dumps of publicly released SNPs were made available via FTP on our website with regular intervals, both in MySQL and Oracle text-table format and various other useful text dumps. This cycle of releases ceased in fall 2001 once the discovery phase was finished, but with the recent additions of genotype and allele frequency data, now available via the DCC FTP-site, a new dump is planned for fall 2002.
ALLELE FREQUENCIES AND GENOTYPES
Recently, the TSC has sponsored several studies to characterize the SNP dataset by genotyping and/or determining allele frequencies in populations. Allele frequencies in populations worldwide have been estimated directly by counting alleles in unrelated, genotyped individuals, and indirectly via pooled methods (7). In a recent pilot study, Gabriel et al. (6) used genotypes from European, Asian and African populations to define haplotype blocks in 51 regions of the genome, demonstrating the usefulness of SNPs in such studies, as well as the presence of large blocks of linkage equilibrium in the human genome.
Allele frequency and genotype data from these studies and others (see next section) have been submitted to the DCC and are available on the DCC website via FTP-download and individual web pages. Table 1 shows a summary of the available data.
SNP LINKAGE MAP
As part of the TSC Linkage Map Project (8), Matise et al. used allele frequency data to select out of an initial set of 6000 TSC SNPs a subset of 2772 that were polymorphic across populations. These were genotyped in 48 CEPH families and used to construct a SNP meiotic linkage map, with an effective map resolution of 4 cM. The data from this study, both the maps themselves and individual genotypes, are available for download via the DCC website. Easy comparison between the SNP linkage map and the genome is made possible by a side-by-side comparative map viewer adapted from the Gramene project (9). This allows researchers to move easily between the linkage map and the corresponding region on the genome. The comparative map viewer continues to be developed in our group and generalized for use in other projects (K. Clark, unpublished).
QUERYING THE DATABASE
On the DCC website, users can query for SNPs by TSC or dbSNP identifiers, or search by genomic location. The results can be restricted to confirmed SNPs, those with allele frequency data, or to those of a certain SNP function class, such as a coding or non-synonomous SNP.
Another popular search function is the gene keyword search which will identify a gene and all nearby SNPs, via a full text search of the gene's name and description. Users can retrieve text table dumps of the results of either of those search functions.
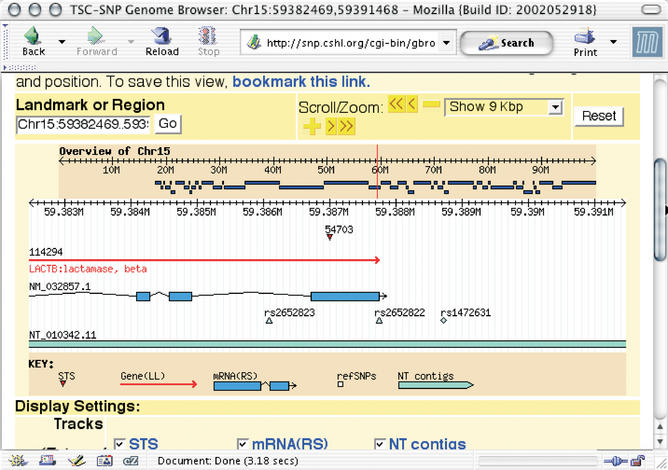
The Gbrowse package (see system design and implementation) allows users to view features on the genome assembly, zoom in and out, pan right or left, customize which features are displayed and their color and more (Fig. 1). Reference SNP clusters, mapped onto the genome assembly by NCBI, which contain submitter SNPs from TSC are hyperlinked to our website, while non-TSC clusters link to dbSNP. Clusters containing TSC SNPs that have allele frequency data associated with them have a special diamond shape, while others have the general triangle shape. These customizations are all done via the GBrowse configuration file and further customizations based on TSC-data are planned.
Figure 1.
A GBrowse display showing part of the gene LACTB on chromosome 15. Three SNPs are shown, one of which has a diamond shape because it has allele frequency data in at least one population.
SYSTEM DESIGN AND IMPLEMENTATION
The TSC repository consists of two subcomponents: (A) a primary Oracle database which contains all TSC SNP data and (B) a secondary MySQL database, serving as the DCC website back end, which contains a subset of the primary database consisting of data for publicly released SNPs. Data used to discover the SNPs, such as tracefiles and related data, are stored on a fileserver, since those are not normally needed after the SNPs have been called, except when website users want to view traces for individual SNPs, in which case the file is retrieved from our FTP-site.
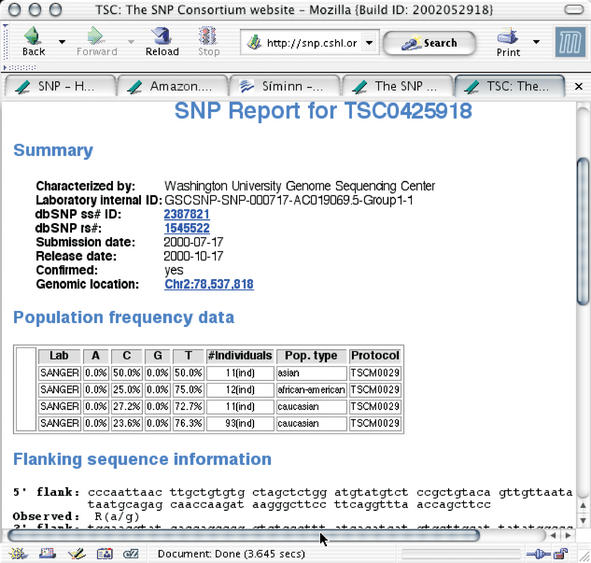
The website software layer consists of a set of simple Perl CGI-scripts that run through the ubiquitous Apache webserver, an HTML-rendering module and a database module. This layer produces webpages for single SNPs (Fig. 2), perform keyword searches for SNP IDs, gene descriptions and more.
Figure 2.
A SNP report showing SNP details, including observed alleles, submitting lab, flanking sequences, genomic location reported allele frequencies and more. Information on this page can be dumped to textfile in various formats, for one or many SNPs at a time.
Recent deployment of the Generic Genome Browser (GBrowse) package (10) from the GMOD project for genome browsing on our website has allowed us to simplify the TSC codebase greatly. GBrowse uses a Bio::DB::GFF database, a subcomponent of the Bioperl toolkit (11), as its back end, which we have loaded with genome annotations available from NCBI. These annotations include genomic coordinates for RefSeq mRNAs, corresponding LocusLink genes, NT contigs, STS markers and, in particular, reference SNP clusters (12). We now use NCBI-produced coordinates for the corresponding reference SNP clusters as the genomic locations for TSC submitter SNPs contained within those clusters. We plan to import into the Bio::DB::GFF database features from Ensembl (3) as well and possibly other sources.
FUTURE DEVELOPMENT
A larger SNP allele frequency dataset was made available for FTP-download in November 2002, containing the full ∼95.000 SNPs characterized in at least one population.
The focus of the website has already shifted from representing SNP discovery data over to a more population centric role. This will undoubtly expand further into the field of human haplotype structure as more data from studies in that field become available. More population-related web page reports are on the horizon, like haplotype feature views, population reports and graphical family pedigree views showing genotypes for multiple SNPs.
It is also important to continue to evolve our database infrastructure and data modeling. We plan to deploy the Extensible Markup Language (XML) technology in the near future, not only as a data-dump format and submission format, but also for internal data handling on top of our relational database system and for data exchange with other repositories. This would facilitate communications with our close collaborator dbSNP.
It is important to note that it is not the intention of the DCC website to compete with much larger projects such as Ensembl (3), the UCSC browser (13) or the NCBI Map Viewer in the realm of whole genome annotation and display. We want to use existing genomic data wherever possible and augment them with our own SNP data, hence the use of the Generic Genome Browser software and NCBI annotations on our website. We intend to continue to focus on the SNP data themselves, on developing customized query and browsing tools and on our role as a data repository, exchanging data and collaborating with dbSNP and others as much as possible. Looking further than this, adopting XML-technology would make it easier for us to offer more general SNP data-sharing WWW-services via tools such as the already well-established Distributed Annotation System (DAS) (14) or the new and rapidly-evolving BioMOBY system. This would enable projects such as Ensembl to use our services in their plug-and-play system, which already supports DAS third-party annotations.
REFERENCES
- 1.Sachidanandam R., Weissman,D., Schmidt,S., Kakol,J., Stein,L., Marth,G., Sherry,S., Mullikin,J., Mortimore,B., Willey,D., Hunt,S., Cole,C., Coggill,P., Rice,C., Ning,Z., Rogers,J., Bentley,D., Kwok,P., Mardis,E., Yeh,R., Schultz,B., Cook,L., Davenport,R., Dante,M., Fulton,L., Hillier,L., Waterston,R., McPherson,J., Gilman,B., Schaffner,S., Van Etten,W., Reich,D., Higgins,J., Daly,M., Blumenstiel,B., Baldwin,J., Stange-Thomann,N., Zody,M., Linton,L., Lander,E. and Altshuler, D. The International SNP Map Working Group (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature, 409, 928–933. [DOI] [PubMed] [Google Scholar]
- 2.Stein L.D. and Thierry-Mieg,J. (1998) Scriptable access to the Caenorhabditis elegans genome sequence and other ACeDB databases. Genome Res., 8, 1308–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hubbard T., Barker,D., Birney,E., Cameron,G., Chen,Y., Clark,L., Cox,T., Cuff,J., Curwen,V., Down,T., Durbin,R., Eyras,E., Gilbert,J., Hammond,M., Huminiecki,L., Kasprzyk,A., Lehvaslaiho,H., Lijnzaadl,P., Melsoppl,C., Mongin,E., Pettett,R., Pocock,M., Potter,S., Rust,A., Schmidt,E., Searle,S., Slater,G., Smith,J., Spooner,W., Stabenau,A., Stalker,J., Stupka,E., Ureta-Vidal,A., Vastrik,I. and Clamp,M. (2002) The Ensembl genome database project. Nucleic Acids Res., 30, 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kent W.J. and Haussler,D. (2001) Assembly of the working draft of the human genome with GigAssembler. Genome Res., 11, 1541–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gabriel S.B., Schaffne,S.F., Nguyen,H., Moore,J.M., Roy,J., Blumenstiel,B., Higgins,J., DeFelice,M., Lochner,A., Faggart,M., Liu-Cordero,S.N., Rotimi,C., Adeyemo,A., Cooper,R., Ward,R., Lander,E.S., Daly,M.J. and Altshuler,D. (2002) The structure of haplotype blocks in the human genome. Science, 296, 2225–2229. [DOI] [PubMed] [Google Scholar]
- 7.Vieux E.F., Kwok,P.Y. and Miller,R.D. (2002) Primer design for PCR and sequencing in high-throughput analysis of SNPs. Biotechniques, 32, 28–30. [PubMed] [Google Scholar]
- 8.Matise T.C., Sachidanandam,R., Kakol,J., Clark,A., Kruglyak,L., Wijsman,E., Buyske,S., Chui,B., Cohen,P., Toma,C., Ehm,M., Glanowski,S., He,C., Heil,J., McMullen,I., Pericak-Vance,M.A., Stein,L.D., Wagner,M., Winick,J., Winn-Deen,E.S., Wilson,A.F., Cann,H.M., Lai,E. and Holden,A.L. A high-resolution human (SNP) linkage map. Nature, in preparation.
- 9.Ware D., Jaiswal,P., Ni,J., Pan,X., Chang,K., Clark,K., Teytelman,L., Schmidt,S., Zhao,W., Cartinhour,S., McCouch,S. and Stein, L. (2002) Gramene: a resource for comparative grass genomics. Nucleic Acids Res., 30, 103–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stein L.D. (2002) The Generic Genome Browser: A building block for a model organism system database. Genome Res., 12, 1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stajich J.E., Block,D., Boulez,K., Brenner,S.E., Chervitz,S., Dagdigian,C., Fuellen,G., Gilbert,J.G.R., Korf,I., Lapp,H., Levaslaiho,H., Matsalla,C., Mungall,C.J., Osborne,B., Pocock,M., Schattner,P., Senger,M., Stein,L.D., Stupka,E., Wilkinson,M.D. and Birney,E. (2002) The Bioperl Toolkit: Perl modules for the life sciences. Genome Res., 12, 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherry S.T., Ward,M.-H., Kholodov,M., Baker,J., Phan,L., Smigielski, E.M. and Sirotkin,K. (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acid Res., 29, 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kent W.J., Sugnet,C.W., Furey,T.S., Roskin,K.M., Pringle,T.H., Zahler,A.M. and Haussler,D. (2002) The human genome browser at UCSC. Genome Res., 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dowell R.D., Jokerst,R.M., Day,A., Eddy,S.R. and Stein,L. (2001) The Distributed Annotation System. BMC Bioinformatics, 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]