Abstract
WormBase (http://www.wormbase.org/) is a web-accessible central data repository for information about Caenorhabditis elegans and related nematodes. The past two years have seen a significant expansion in the biological scope of WormBase, including the integration of large-scale, genome-wide data sets, the inclusion of genome sequence and gene predictions from related species and active literature curation. This expansion of data has also driven the development and refinement of user interfaces and operability, including a new Genome Browser, new searches and facilities for data access and the inclusion of extensive documentation. These advances have expanded WormBase beyond the obvious target audience of C. elegans researchers, to include researchers wishing to explore problems in functional and comparative genomics within the context of a powerful genetic system.
DESCRIPTION
Caenorhabditis elegans is a soil nematode whose small size (1 mm), fast generation time (3 days), ease of culturing, and the ability to maintain strains by either clonal or sexual reproduction have all contributed to its widespread use as a genetic model organism. Furthermore, C. elegans is transparent, exhibits an invariant cell lineage and has a relatively simple nervous system, facilitating studies of development and nervous system function. Finally, its small genome size and gene complement (100 Mbp, 19 473 genes) and complete genome sequence have extended the benefits of C. elegans to studies in genomics and proteomics (1).
WormBase is a collaborative project whose aim is to consolidate the considerable information on the biology of C. elegans (2). In particular, we seek to provide access to this data in a user-friendly format without compromising the power and flexibility of more advanced queries. As an advanced genetic model organism database, WormBase contains substantial information in those areas that helped establish the worm as a model organism. These data include: (i) the essentially complete genome sequence (3); (ii) the developmental lineage of the worm (4,5); (iii) the connectivity of the nervous system (6); (iv) mutant phenotypes, genetic markers and genetic map information; (v) gene expression described at the level of single cells; and (vi) bibliographic resources including paper abstracts and author contact information.
WormBase continues to emphasize curation of the central information infrastructure, while expanding the biological scope of the resource. Current objectives include systematic curation of the C. elegans literature, the integration of large-scale, community submitted datasets and the development of simplified methods of data access. A survey of some of the new features of the WormBase resource are described below.
Genome Browser
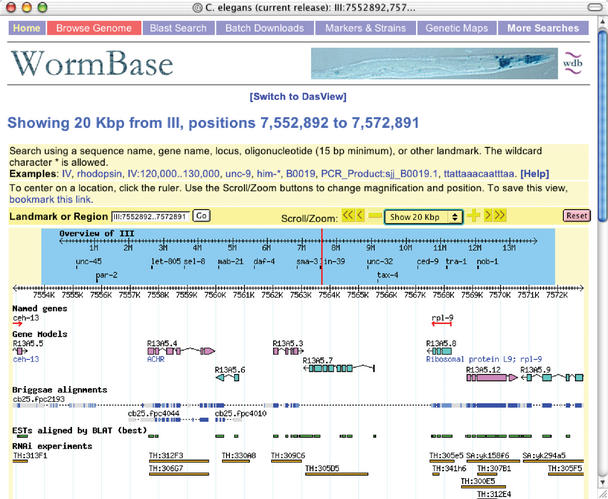
Of the substantial additions and refinements to the user interface, the most apparent is the implementation of a new Genome Browser (7). This Genome Browser features a highly configurable interface, preservation of user preferences, semantic zooming, an extensible plug-in based architecture and the ability to display user annotations within the context of the C. elegans genome. Users can enter the Genome Browser through hypertext links from related report pages, or can search from the Genome Browser interface directly using a marker name or position, chromosomal coordinates, or a description of biological function. In instances where multiple items are returned, a selection display is presented showing the position of items returned in the genome. Selecting an individual item takes the user into the graphical representation of that region of the genome (Fig. 1).
Figure 1.
An expanded view of the Genome Browser, display a variety of features including genes, alignments with C. briggsae, ESTs and RNAi experiments.
The Genome Browser was developed as a central component of the Generic Model Organism Database (GMOD) project, an effort to develop extensible and reusable components for model organism databases. More information on the Genome Browser and the software that drives it can be found at http://www.gmod.org/.
Caenorhabditis briggsae sequence and gene predictions
In addition to maintaining information on C. elegans, WormBase now contains the essentially complete genomic sequence from the related nematode C. briggsae. This information will be useful for identifying conserved regions between the two genomes, for verifying and correcting gene models, for identifying gene family expansion and contractions, and for studying the functional differences between specific proteins.
The C. briggsae genome is browsable and searchable in both the C. elegans Genome Browser and in a separate instance that displays C. briggsae-specific analysis. In the C. elegans Genome Browser, nucleotide-level alignments to C. briggsae calculated using the Blat (8) and WABA (9) algorithms are displayed (Fig. 1). The C. briggsae specific Genome Browser displays gene predictions from a number of ab initio gene prediction algorithms and full WABA and BLATX alignments to C. elegans. Raw analysis files of the C. briggsae genome can be retrieved from the WormBase FTP site (ftp.wormbase.org).
Single nucleotide polymorphisms (SNPs)
WormBase now includes 6386 SNPs (10). These SNPs have been of great utility to the research community, accelerating the pace of genetic mapping and increasing its resolution. To facilitate the identification of SNPs and genetic markers suitable for mapping experiments, two new searches have been developed. Provided with a genetic or physical interval, these searches return all genetic and physical markers contained within the interval. A basic search provides quick access to this information. A more extensible version of this search allows a user to restrict returned markers based on their ease of scoring, lethality to the organism, and, in the case of SNPs, on whether they generate a restriction fragment length polymorphism. Freely-available strains carrying genetic markers are also listed, simplifying the acquisition of experimental reagents.
RNAi
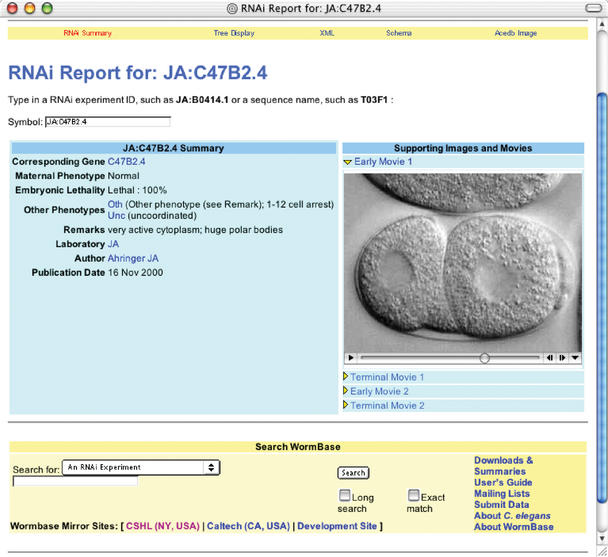
Results from systematic RNA interference screens as well as individual experiments are now available for 7242 genes. These data are accessible in two ways: first, they can be searched by phenotype through a specific RNAi search page; second they are presented on the descriptive pages for individual genes (Fig. 2). Thus, users searching for candidate genes that result in specific phenotypes when disrupted as well as users studying the function of individual genes have direct access to the data. Results of experiments include various descriptive terms of the mutant phenotype, the span of the gene segment targeted, and in many cases, QuickTime movies displaying the resulting phenotype.
Figure 1.
A typical page for an RNAi experiment. QuickTime movies of many results help to illustrate developmental timing defects.
Expression patterns and profiles
In addition to information on reporter gene constructs, WormBase now presents data for consolidated microarray experiments from a variety of life stages and conditions (11). In the analysis of these experiements, genes are grouped based on their tissue or life-stage expression profile; genes expressed in similar tissues coalesce into a mountain when displayed in 3D topology. WormBase displays these microarray profiles in a 2D representation, allowing users to see which mountain a particular gene of interest belongs, as well as identify genes likely to be coexpressed based on their distance from that mountain in the display.
Neuron searches
Although there is substantial information on the anatomy of the worm available, this information contains a level of abstraction that makes it difficult to relate to the organism as a whole. In order to make this data more accessible, WormBase has implemented neuron-specific search pages. These searches enable users to search for neurons of specific classes. For example, a user can search for all neurons derived from a specific lineage, or search for all GABA-releasing neurons.
Gene verification and model correction
Of major interest to end-users is the integrity and accuracy of gene predictions. WormBase is using a variety of methods to address difficulties in gene prediction. First, data from the C. elegans ORFeome project has been integrated into the WormBase infrastructure. By systematically amplifying genes from a cDNA library, the ORFeome project sought to define all transcribed genes and correct their splicing patterns (12). These experiments, in the form of PCR primer pairs and amplified products, are displayed on the Genome Browser allowing users to quickly determine if a predicted gene was amplified by the technique. Second, gene models have been enhanced by the annotation and display of UTRs on the Genome Browser. Finally, gene models continue to be revised from user submissions and literature curation and a new gene representation in the database schema is being implemented to track revisions in gene structure.
Gene Ontology
The Gene Ontology project is an attempt to develop a controlled vocabulary to describe biological processes and molecular functions (http://www.geneontology.org/) (13). WormBase staff work in close collaboration with the Gene Ontology project. Currently, 11 593 terms have been assigned and a new Gene Ontology browser makes it easy to search for genes associated with a specific ontology term. For example, a search for Map Kinase displays all genes associated with the Map Kinase function. Results are displayed in a hierarchical format, allowing the end user to easily explore the Ontology structure by selecting terms above or below their gene of interest.
Curation and annotation
Systematic literature curation continues to play an important role in expanding the biological scope of WormBase and in assessing the data integrity of the database. A first pass curation step outlines the types of data contained in papers. Second pass curation now focuses on gene structure, function and expression information. Detailed gene function summaries are also being generated to place this data into an integrated context.
FACILITATING DATA ACCESS
WormBase provides access to data via multiple methods and in multiple formats. Primary access to WormBase is provided through the web interface. Data from any single display is available in XML format, and new interfaces provide batch access to data. A ‘Batch Info’ form gives users a convenient utility to compare and retrieve a variety of information about sets of genes. This information includes relevant sequences, mutant phenotypes, prominent motifs and blast homologies, as well as identifiers for external databases. A flexible Genome Dumper allows users to retrieve any type of sequence feature or region, and to do this in a manner relative to other sequence features. For example, users can request a specified quantity of sequence upstream from every gene.
For users interested in programmatic access to the resource, WormBase provides scriptable access to the underlying ACeDB database via the AcePerl module (stein.cshl.org/aceperl/). Furthermore, a growing number of common precomputed data sets, such as spliced, unspliced and translated sequences of predicted and confirmed genes, tRNAs, C. briggsae sequences, gene predictions, and alignments as well as the software that drives WormBase, are available through the WormBase FTP site (ftp.wormbase.org).
FUTURE DIRECTIONS
WormBase is an ongoing project. A number of new features are currently in development to expand the scope and usability of the resource.
Large scale data-sets and literature curation
Among large scale data-sets currently under curation or in planning stages are two hybrid protein-interactions (14), additional verified single nucleotide polymorphisms (15) and the assignment of orthologs and paralogs across species. Literature curation is now being expanded to included cis and trans gene-regulation and gene–gene interactions.
Tools for comparative genomics
Currently, we are working to expand the tools and available data to assist in comparative genomics analyses. This includes integrating information on orthology, paralogy and genome rearrangements between C. briggsae and C. elegans. These data will be integrated into Genome Browser displays, as well as side-by-side comparisons with C. elegans on a gene-by-gene level.
Enhanced user interface and access
We continue to refine the user interface of WormBase to present diverse data types in both biologically and experimentally relevant context. A more pliable user interface, which allows users to select the types of resources that they most frequently access is currently under development. With the increasing complexity of the resource, the WormBase consortium has initiated extensive documentation efforts, opening the resource to researchers outside of the C. elegans community. Tools to enhance curation are also under development, such as scripts to simplify the submission of data from the community. Finally, with the growing focus on genome-wide screens, we aim to provide streamlined access to large quantities of data for both biologists and bioinformaticists through highly customizable searches.
Acknowledgments
ACKNOWLEDGEMENTS
WormBase is supported by grant P41-HG02223 from the US National Human Genome Research Institute and the British Medical Research Council. P.W.S. is an investigator with the Howard Hughes Medical Institute.
REFERENCES
- 1.Riddle D.L., Blumenthal,T., Meyer,B.J. and Priess,J.R. (1997) C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 2.Stein L., Sternberg,P., Durbin,R., Thierry-Mieg,J. and Spieth,J. (2001). WormBase: network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res., 29, 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The C. elegans Genome Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 4.Sulston J.E., Schierenberg,E., White,J.G. and Thomson,J.N. (1983) The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol., 100, 64–119. [DOI] [PubMed] [Google Scholar]
- 5.Sulston J. and Horvitz,H.R. (1977) Post-embryonic cell lineages of the nematode Caenorhabditis elegans. Dev. Biol., 56, 110–156. [DOI] [PubMed] [Google Scholar]
- 6.White J.G., Southgate,E., Thomson,J.N. and Brenner,S. (1986) The structure of the nervous system of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond., 314, 1–340. [DOI] [PubMed] [Google Scholar]
- 7.Stein L., Mungall,C., Shu,S., Caudy,M., Mangone,M., Day,A., Nickerson,E., Stajich,J., Harris,T.W., Arva,A. et al. (2002) The Generic Genome Browser: a building block for a model organism system database. Genome Res., 12, 1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kent W.J. (2002) BLAT—the BLAST-like alignment tool. Genome Res., 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kent W.J. and Zahler,A.M. (2000) Conservation, regulation, synteny, and introns in a large-scale C. briggsae–C. elegans genomic alignment. Genome Res., 10, 1115–1125. [DOI] [PubMed] [Google Scholar]
- 10.Wicks S.R., Yeh,R.T., Gish,W.R., Waterston,R.H. and Plasterk,R.H. (2001) Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nature Genet., 28, 160–164. [DOI] [PubMed] [Google Scholar]
- 11.Kim S.K., Lund,J., Kiraly,M., Duke,K., Jiang,M., Stuart,J.M., Eizinger,A., Wylie,B.N. and Davidson,G.S. (2001) A gene expression map for Caenorhabditis elegans. Science, 293, 2087–2092. [DOI] [PubMed] [Google Scholar]
- 12.Reboul J., Vaglio,P., Tzellas,N., Thierry-Mieg,N., Moore,T., Jackson,C., Shin-i,T., Kohara,Y., Thierry-Mieg,D., Thierry-Mieg,J. et al. (2001) Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nature Genet., 27, 332–336. [DOI] [PubMed] [Google Scholar]
- 13.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T. et al. (2000) Gene ontology: tool for the unification of biology. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walhout A.J., Sordella,R., Lu,X., Hartley,J.L., Temple,G.F., Brasch,M.A., Thierry-Mieg,N. and Vidal,M. (2000) Protein interaction mapping in C. elegans using proteins involved in vulval development. Science, 287, 116–122. [DOI] [PubMed] [Google Scholar]
- 15.Swan K.A., Curtis,D.E., McKusick,K.B., Voinov,A.V., Mapa,F.A. and Cancilla,M.R. (2002) High-throughput gene mapping in Caenorhabditis elegans. Genome Res., 12, 1100–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]