Abstract
GrainGenes, http://www.graingenes.org, is the international database for the wheat, barley, rye and oat genomes. For these species it is the primary repository for information about genetic maps, mapping probes and primers, genes, alleles and QTLs. Documentation includes such data as primer sequences, polymorphism descriptions, genotype and trait scoring data, experimental protocols used, and photographs of marker polymorphisms, disease symptoms and mutant phenotypes. These data, curated with the help of many members of the research community, are integrated with sequence and bibliographic records selected from external databases and results of BLAST searches of the ESTs. Records are linked to corresponding records in other important databases, e.g. Gramene's EST homologies to rice BAC/PACs, TIGR's Gene Indices and GenBank. In addition to this information within the GrainGenes database itself, the GrainGenes homepage at http://wheat.pw.usda.gov provides many other community resources including publications (the annual newsletters for wheat, barley and oat, monographs and articles), individual datasets (mapping and QTL studies, polymorphism surveys, variety performance evaluations), specialized databases (Triticeae repeat sequences, EST unigene sets) and pages to facilitate coordination of cooperative research efforts in specific areas such as SNP development, EST-SSRs and taxonomy. The goal is to serve as a central point for obtaining and contributing information about the genetics and biology of these cereal crops.
DESCRIPTION
Data content
GrainGenes was created in 1992 by the US Department of Agriculture to facilitate distribution of data needed by small-grains breeders, pathologists, geneticists and molecular biologists to create improved cultivars of Triticeae (wheat, barley, rye, triticale) and oat crops.
A major focus of interest for all these groups of researchers is genetic mapping of molecular markers, genes and quantitative traits. GrainGenes provides information about these mapping studies in as much depth as can be obtained, more than is feasible in print publications. The additional data are submitted directly by the authors and include such details as raw mapping scores, autoradiograms labeled to show which polymorphic fragment corresponds to which locus, characteristics of cloned RFLP probes, PCR primers and reaction conditions and statistical parameters from quantitative trait locus (QTL) analysis. This information is being used to transfer markers to new maps in different laboratories, add new markers to existing maps, and build consensus maps from separate map studies (1,2). Genetic mapping continues to be an active area of research for these crops, as the advent of highly polymorphic markers like AFLPs and SSRs has permitted mapping directly in breeding lines of interest (3).
Currently GrainGenes contains 81 full-genome maps, accessed under the Map_Data category at www.graingenes.org. Many more have been published and are in the pipeline to add to the database.
Another major component of GrainGenes is information about genes identified in the small-grain crops. GrainGenes serves as the database-structured form of the compendia (4–6) summarizing decades of classical genetics of traits affecting morphology, resistance to pests and stresses, phenology (flowering time etc.), isozymes and storage proteins.
Information about these classical genes includes
accepted names and synonyms,
map locations,
alleles, with their phenotype descriptions, genetic stocks and occurrence in cultivars and other germplasm,
key references, and
images of naked-eye polymorphisms and disease symptoms.
The most complex data structure in GrainGenes is the one describing QTL studies, due to the multivariate nature of such experiments (trait×genotype×environment). The central data class here is the Trait_Study, which comprises the information about one trait in one mapping population in a set of environments. From the Trait_Study record are links to the Environment records, a Trait_scores record for each environment containing the values for all members of the population, QTL records for the QTLs reported, and a single Map_Data record for the molecular marker scores of the population. Each QTL links to its Map, and most classes link to each other for convenience. The overarching data class is the Reference, which usually encompasses Trait_Studies for several traits. The ease with which data of this complexity can be structured in a natural way derives in large part from the flexibility of the underlying ACEDB software.
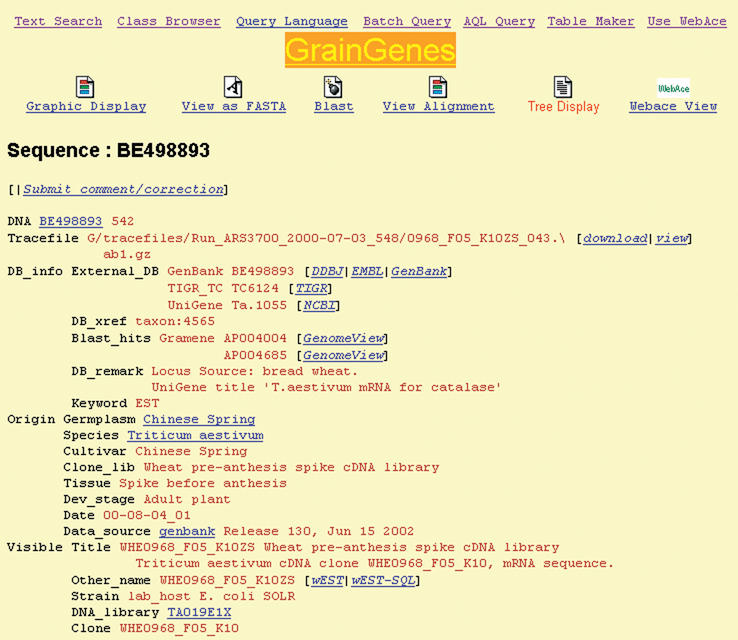
New Triticeae research areas are opening rapidly due to large-scale EST sequencing of these species. As of August 2002, there are over 450 000 wheat and barley ESTs in the dbEST database, more than any single organism except human and mouse and another 500 000 are expected in the next 18 months. The number of sequence records in GrainGenes nearly tripled in the last year. Information about these ESTs (Fig. 1) includes the GenBank record, top BLAST sequence similarities, contig assembly and connections to external databases like the TIGR Gene Indices, NCBI UniGenes and Gramene homologies to the rice genome. Connections are also made to the in-house relational database ‘wEST’, containing detailed BLAST results and mapping data for ESTs from the International Triticeae EST Cooperative (http://wheat.pw.usda.gov/genome) and the NSF-supported US wheat EST project (http://wheat.pw.usda.gov/NSF). EST-based data being generated now that will be flowing into GrainGenes include EST-SSRs, SNPs, cross-species mapping and gene expression.
Figure 1.
A sample EST sequence record from GrainGenes.
An independent but closely related database that is important for many GrainGenes users is Gramene (7). The focus of Gramene is inter-species comparisons amongst the grasses (Poaceae), especially versus rice. It also includes detailed information about rice maps, sequences, traits, etc. much as GrainGenes does for Triticeae and Avena species. Interconnections between the two databases are supplied to facilitate moving back and forth.
Navigating GrainGenes
The GrainGenes Database is designed to make information retrieval by browsing as easy as possible, to start from any entry point such as a gene or map or trait and find any related data of interest by ‘following one's nose’ from record to record. This is possible in part due to features of the software, in part by structuring the data appropriately. However, there are some constraints, so the path from entry to target cannot always be completely intuitive. For example, Gene records do not connect directly to Maps, but indirectly via Locus records.
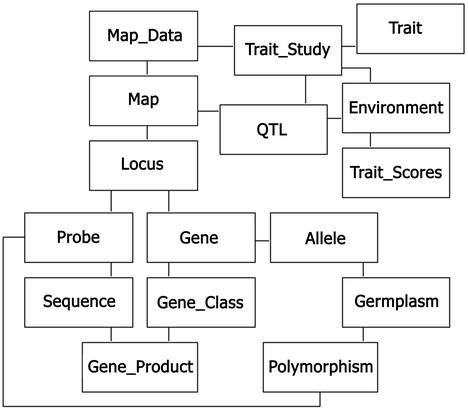
A map of the primary data paths in GrainGenes is given in Figure 2. The connections indicated are curated rigorously, so this map may be used to ascertain whether some desired information is in the database or not. For example, if a particular gene has no Locus link, it is not on any of the maps in the database. Many additional links exist for convenience but cannot be relied upon in this way. All the connections in Figure 2 are bidirectional but some others are not. e.g. all Sequences link to the Species from which they were derived, but connecting ‘Triticum aestivum’ to all 180 000 sequences derived from it would not be helpful for browsing and would degrade performance of the database server. The information is still available by querying (below), in this case ‘find sequence species=T. aestivum’.
Figure 2.
Recommended browse map of GrainGenes. Secondary interconnections and some additional data classes are omitted for clarity.
A graphical display mode is available for records of class Map, Locus, QTL and Sequence, and is the default mode for the former three. Buttons ‘Graphic Display’ and ‘Text Display’ at the top of the web page switch between modes.
Synonymies
Nomenclatural variations in Gene, Allele, Probe, Species and Germplasm are managed by establishing one canonical name for each entity. Synonyms are in separate data records that link only to the canonical record. All connections to related data (Fig. 2) are via the canonical record. The Locus class is treated differently: each map is displayed using the locus names as given in the published reference rather than trying to impose a naming rule. Loci mapped with the probe BCD98, for example, have been published under 15 names—BCD98, Xbcd98-1, Xcnl.BCD98, Xbcd98-1A.1, et al. To find all the potentially orthologous loci, use the controlled class Probe as the starting point for browsing or querying. Loci for mapped genes are handled similarly using the controlled class Gene; such loci are usually connected to Gene_Class too, since in some cases it is uncertain which gene, as defined functionally, corresponds to a locus mapped using the cloned gene as a molecular marker.
Query access to GrainGenes
Like most other databases, GrainGenes access modes include simple text searches and boolean queries, producing a list of records to choose from. Extensions and additional modes include, first, a feature of the basic query language that allows a query to navigate from a given record to other linked records as described above for browsing. For example ‘find Gene glu*; follow Locus; follow Map’ returns a menu of all maps containing Glu (glutenin) genes. Second, the query language also can operate batchwise, accepting a pasted list of query terms instead of a single argument (www.graingenes.org/cgi-bin/ace/custom/bquery/graingenes). Finally, two query interfaces allow users to extract tabular output of any desired fields in the database, whether a complete dump or restricted by conditions on those or related fields. AQL Query uses an SQL-like syntax, whereas TableMaker provides column by column query builder assistance. Both are useful for exporting GrainGenes data into spreadsheets or local databases, and both accept parameterized input from web forms like those on the GrainGenes Quick Queries page (http://wheat.pw.usda.gov/ggpages/ggtabledefs.html), so experienced users can build arbitrarily powerful query forms.
Other access points
Besides the primary server at www.graingenes.org the database is online at mirror sites in California, Canada, UK, Belgium, France and Japan (http://wheat.pw.usda.gov/ggpages/pickGG.shtml). It can be downloaded in ACEDB format from ftp://ftp.graingenes.org or ftp://grain.jouy.inra.fr/pub/database/, both updated daily. Direct aceclient access is available on request. Selected maps exported from GrainGenes are available in NCBI's Plant Genomes Central, Gramene and BarleyDB.
GrainGenes home page
The GrainGenes Database described above is only one item on the GrainGenes WWW home page, http://wheat.pw.usda.gov. The top page itself is a frequently updated gallery of links to ‘Hot Topics’ of interest. The Publications page contains a large amount of full-text information, including the annual newsletters for wheat, barley and oat, as well as monographs and individual articles. Much of this textual data was converted from hard copy to electronic form by the GrainGenes project and most of it is not available online elsewhere. The Employment page is quite active, with new postings almost every day. There are pages for five primary research categories—Genomics, Mapping, Germplasm, Pathology and Taxonomy—containing individual datasets (mapping and QTL studies, polymorphism surveys, variety performance evaluations), links that query for specific datasets from the GrainGenes Database, and pointers to external sites of interest.
Particularly important sections of the GrainGenes website are those dedicated to coordinating community research efforts in specific areas, e.g. SNP development, EST-SSRs and taxonomy. These web pages include not only general information about the projects but downloadable data, targeted BLAST datasets and discussions among the participants, backed up with special-focus email newsgroups. Some of the currently active ones are:
TREP, the Triticeae Repeat Sequence Database (http://wheat.pw.usda.gov/ITMI/Repeats/), curated by Thomas Wicker. An annotated, BLASTable collection of repetitive DNA sequences from wheat, barley, rye and related species.
Wheat SNP Development (http://wheat.pw.usda.gov/ITMI/2002/WheatSNP.html), organized by Peter Isaac. A baseline assembly of 186 000 ESTs has been built and large contigs are being assigned to specific research groups to identify and validate SNPs that distinguish alleles as well as the three homoeologous genes expected in bread wheat (hexaploid).
Triticeae EST-SSR Coordination (http://wheat.pw.usda.gov/ggpages/ITMI/2002/EST-SSR), organized by Nils Stein. Microsatellite-containing ESTs from grass (Poaceae) species, assembled into non-redundant ‘Uni-EST-SSR’ contigs, with identification of cross-species clusters. These are being tested by several laboratories for usefulness as markers for mapping and genetic diversity assessment.
DISCUSSION
GrainGenes could not have been created and cannot be maintained without the active participation of the many scientists who are providing and using the information and the research progress underlying it. Major curators of large datasets at the beginning were Gary Hart (Texas A&M U., US) for wheat genes from the Catalogue of Gene Symbols for Wheat, and Ken Kephart (Montana State U., US) for data about germplasm, pathology and taxonomy. Currently, Udda Lundqvist (Nordic Gene Bank, SE) is preparing the definitive data on barley genes (www.untamo.net/bgs/) and Rudi Appels (Murdoch U., AU) is building a composite map of wheat. The many other GrainGenes contributors over the years have been acknowledged piecemeal on the What's New pages (http://wheat.pw.usda.gov/ggpages/whatsnew/), but we wish to take this opportunity to thank them all: please see the Supplementary Material. Suggestions, corrections, bug reports and requests for special queries are always welcome. Please write to curator@wheat.pw.usda.gov.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
GrainGenes is a product of the US Department of Agriculture, Agricultural Research Service, CRIS 5325 21000 007 00D. Our mirror sites worldwide have been invaluable in maintaining uninterruptible GrainGenes Database accessibility regardless of occasional hardware or network problems, especially the INRA site http://grain.jouy.inra.fr which hosts a copy of the GrainGenes WWW site as well. Important software support is being provided by Jean Thierry-Mieg for ACEDB and Lincoln Stein for AceBrowser. The public data resources Gramene, NCBI and TIGR have been active in assisting export of their data into GrainGenes. We thank Philippe Leroy for suggesting the need for an organizational map of the database, and Mark Sorrells and Tim Close for many helpful discussions.
REFERENCES
- 1.Langridge P., Karakousis,A., Collins,N., Kretschmer,J. and Manning,S. (1995) A consensus linkage map of barley. Mol. Breed., 1, 389–395. [Google Scholar]
- 2.Qi X., Stam,P. and Lindhout,P. (1996) Comparison and integration of four barley genetic maps. Genome, 39, 379–394. [DOI] [PubMed] [Google Scholar]
- 3.Langridge P., Lagudah,E.S., Holton,T.A., Appels,R., Sharp,P.J. and Chalmers,K.J. (2001) Trends in genetic and genome analyses in wheat: a review. Aust. J. Agric. Res., 52, 1043–1077. [Google Scholar]
- 4.McIntosh R.A., Hart,G.E. and Gale,M.D. (1995) Catalogue of gene symbols for wheat. In Li,Z.S. and Xin,Z.Y. (eds), Proceedings of the Eighth International Wheat Genetics Symposium. China Agricultural Scientech Press, pp. 1333–1500.
- 5.Lundqvist U., Franckowiak,J.D. and Konishi,T. (1997) Barley genetic stocks, new and revised descriptions. Barley Genet. Newsl., 26, 44–516. [Google Scholar]
- 6.Marshall H.G. and Shaner,G.E. (1992) Genetics and inheritance in oat. In Marshall,H.G. and Sorrells,M.E. (eds), Oat Science and Technology. ASA, CSSA, SSSA, Madison, WI, vol. 33, 509–571. [Google Scholar]
- 7.Ware D., Jaiswal,P., Ni,J., Pan,X., Chang,K., Clark,K., Teytelman,L., Schmidt,S., Zhao,W., Cartinhour,S., McCouch,S. and Stein,L. (2002) Gramene: a resource for comparative grass genomics. Nucleic Acids Res., 30, 103–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.