Abstract
Rice is not only a major food staple for the world's population but it also is a model species for a major group of flowering plants, the monocotyledonous plants. Draft genomic sequence of two subspecies of rice, Oryza sativa spp. japonica and indica ssp. are publicly available. To provide the community with a resource to data-mine the rice genome, we have constructed an annotation resource for rice (http://www.tigr.org/tdb/e2k1/osa1/). In this resource, we have annotated the rice genome for gene content, identified motifs/domains within the predicted genes, constructed a rice repeat database, identified related sequences in other plant species, and identified syntenic sequences between rice and maize. All of the data is available through web-based interfaces, FTP downloads, and a Distributed Annotation System.
INTRODUCTION
Although there are varying criteria that can be invoked in the selection of a species for genome sequencing, for rice, the merits of sequencing the genome are many. Not only is rice itself the major caloric food source for the world's population (1), but rice also is a central lynchpin for comparative studies in the Gramineae family (2) which contains an overwhelming number of agriculturally relevant crop species including maize, wheat, barley, oat, sorghum, and sugarcane. For these reasons, rice has been the focus of not one, but four, genome sequencing efforts (3–6). From these four endeavors, two public and two private, draft sequence is available for two subspecies of Oryza sativa providing rich resources for not only dissecting the rice genome but also performing comparative studies between two subspecies of rice and between rice and other cereal species.
To provide a robust and centralized resource for rice, we have performed a range of bioinformatic analyses on the public rice genome sequence data generated by the International Rice Genome Sequencing Project (IRGSP). These analyses include anchoring publicly available rice bacterial artificial chromosome and P1 artificial chromosome clones (BAC, PAC) to the genetic map, annotation of BAC/PAC sequences, classification of domains and motifs within proteins predicted in the genome, construction of a rice repeat database, and identification of related sequences in other plant species. These analyses allow immediate access to the rice genome and a platform to data-mine the genome of the world's most important plant.
Anchoring of the rice BAC/PAC clones to the chromosomes
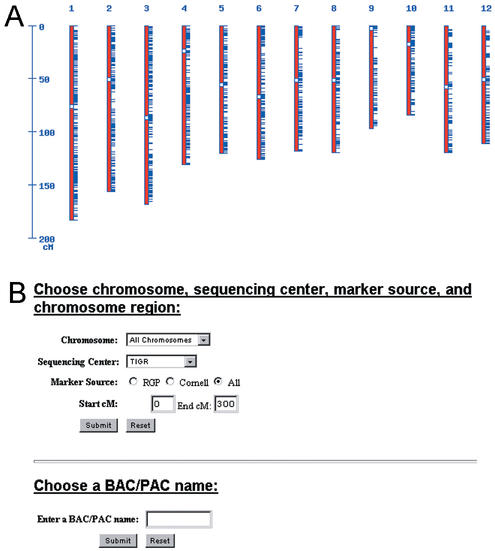
In the public IRGSP sequencing effort, BAC or PACs are sequenced and released to the public databases. Although chromosomal location is provided by the sequencing center with respect to the 12 rice chromosomes, the precise position of the BAC/PAC on the chromosome is not typically included in these submissions. To link the genetic map with the genome sequence, we perform robust in silico alignments of the rice BAC/PACs with all available sequenced genetic markers (http://www.tigr.org/tdb/e2k1/osa1/BACmapping/description.shtml; Fig. 1). A total of 13 896 marker sequences are aligned with the rice BAC/PAC sequences using high stringency cutoff criteria. Currently, we have anchored 2585 (359 Mb) of the 2910 available rice BAC/PAC sequences (401 Mb) to a marker sequence. These alignments provide a robust resource for positional cloning of genes in rice and can be viewed through web displays of each of the chromosomes or through search tools that provide selection based on BAC/PAC name, chromosome, sequencing center, marker source or cM position (Fig. 1).
Figure 1.
In silico alignments of the rice BAC/PAC sequences with rice genetic markers. (A) Clickable display of the 12 rice chromosomes (red) with anchored BAC/PAC clones (blue ticks). (B) Selection of chromosome, sequencing center, marker source, cM position and BAC/PAC name.
Annotation of rice sequences
Finished, phase 2, and phase 3 rice BAC/PAC sequences were downloaded from the PLN and HTGS divisions of GenBank and loaded into osa1, a Sybase relational database. These sequences were annotated for gene content using an automated set of processes that involves ab initio gene finders and database searches against plant nucleic acid and protein databases. The rice sequences were processed with multiple ab initio gene finders including FGENESH (http://www.softberry.com), Genemark.hmm (rice matrix; http://opal.biology.gatech.edu/GeneMark/eukhmm.cgi), Genscan (maize matrix; http://genes.mit.edu/GENSCAN.html), Genscan+ (Arabidopsis matrix; http://genes.mit.edu/GENSCAN.html) and GlimmerM (rice matrix; http://www.tigr.org/tdb/glimmerm/glmr_form.html). Database searches were performed with the TIGR plant gene indices that represent clustered assemblies of EST sequences (7) from multiple plant species including rice, maize, wheat, barley, rye, cotton, sorghum, Arabidopsis, tomato, potato, soybean, ice plant, and Medicago. Database searches were also performed using a modified non-redundant protein database that includes only plant sequences. The output from the gene prediction programs and database searches were stored in osa1. Working models were generated using the FGENESH output and putative identification for the gene was obtained from the most significant database match while models with no significant database match were labeled as hypothetical proteins. Transfer-RNAs are identified using tRNA-Scan SE (8).
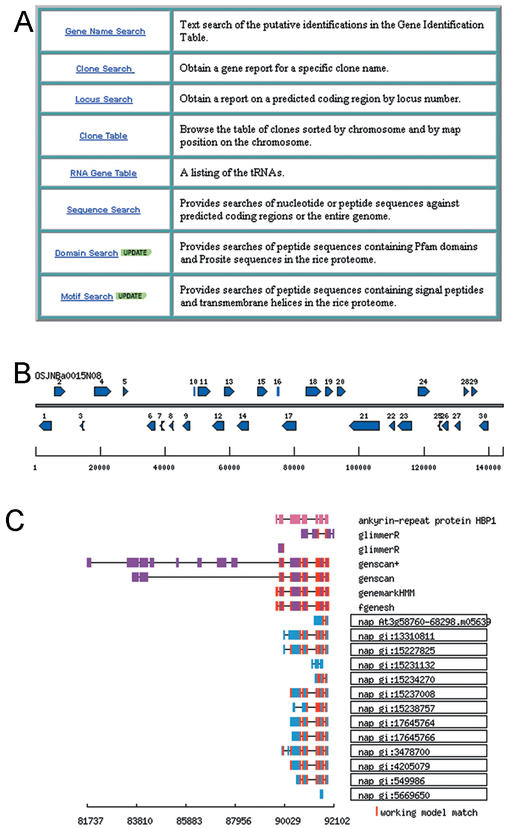
The annotation data is available through a web-based interface in which options are provided for the user to view the annotation data. As shown in Figure 2, tools are available to search the annotation data by specific gene name, BAC/PAC clone name, locus name, or by chromosome location. Once a BAC/PAC clone is selected, working models, along with the putative identification, are displayed in a graphical format for the user (Fig. 2). Additional detail on each model, including gene prediction program output and database search evidence, can be selected by the user (Fig. 2). An example of the gene prediction program output and the database evidence for a single gene model is shown in Figure 2. The annotation data is also available through a Distributed Annotation System (9; Fig. 3; http://www.tigr.org/tdb/e2k1/osa1/irgsp/das.shtml) as well as through FTP download of flatfiles (ftp://ftp.tigr.org/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/).
Figure 2.
Access to annotation data in the TIGR Rice Genome Annotation Resource. (A) Selection tools available to the user for annotation data. (B) Graphic display of gene models on chromosome 3 rice BAC OSJNBa0015N08. (C) Display of database evidence and gene prediction program output for gene model #19 that encodes a putative ankyrin repeat protein HBP1.
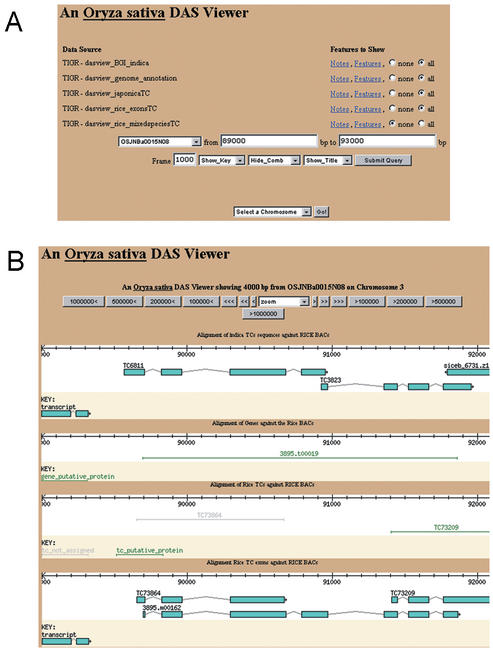
Figure 3.
Distributed annotation system for rice. (A) Selection of views for the TIGR Oryza sativa DAS Viewer. Users can select alignments of the IRGSP rice BACs with the BGI indica ESTs, TIGR rice genome annotation, japonica ESTs, exon-level displays with the genome annotation and japonica ESTs, or TCs from other plant species. Selection tools are also available for clone, chromosome, and fragment length. (B) Alignment of working model # 19 from the chromosome 3 BAC OSJNBa0015N08 with the DAS viewer.
The current version of osa1 contains 369 Mb of rice genomic DNA from 2668 BACs/PACs from the on-going public IRGSP sequencing effort. As the rice genome is estimated to be ∼430 Mb (10) and assuming an overlap of 10% on average between BAC/PAC clones this represents 77% of the rice genome. We have been able to identify a total of 57 442 genes of which we were able to assign a putative function to ∼70%. An average rice gene is 2.51 kb in length and is distributed every 6.4 kb in the genome. The predicted genes in osa1 are further annotated through the identification of motifs and domains which are valuable in assigning function. We have identified signal peptides, uncleaved signal anchors, transmembrane domains, ProSite pattern/profiles and Pfam domains/families in the proteins predicted within the rice genome using the motif/domain finding algorithms SignalP (11) and TMHMM V2.0 (12) and by searching the Pfam (13) and ProSite (14) databases. These data can be queried through a web-based interface by selecting chromosome, sequencing group, or BAC/PAC clone name (http://www.tigr.org/tigr-scripts/e2k1/irgsp.spl).
Classification of repetitive sequences in rice
Repetitive sequences have been documented in rice and include transposable elements, centromere-related sequences, telomere-related sequences, rRNA genes, and other unclassified repetitive sequences. Some of these have known biological functions, e.g., rRNA genes, centromeres and telomeres, whereas the function of the remaining repetitive sequences is unknown. Using known repeat sequences from rice and other cereal species, we have generated a repeat database for rice that contains 19 074 sequences representing 7.6 Mb of sequence. This database is available for BLAST searches (http://www.tigr.org/tdb/e2k1/osa1/blastsearch.shtml) and FTP download (ftp://ftp.tigr.org/pub/data/o_sativa/osa1/PUBLICATION_RELEASE/TIGR_Rice_Repeats).
Identification of related sequences in other plant species
Sequencing the rice genome is not of interest solely to rice biologists. In fact, due to its collinearity with other cereals (2) and appealing features as a model for monocotyledonous plants (15), it is of interest to non-rice plant biologists as well. Thus, it is critical to generate resources to leverage the rice genome and thereby maximize the gain from obtaining the sequence of this species. Through web-based interfaces, we provide three levels of alignments of the rice genome with other plant genomic sequences: low stringency alignments, high stringency alignments, and syntenic alignments. These three levels allow for alternative views of sequence conservation across the plant kingdom and provide multiple entry points for data-mining the rice genome for orthologues and paralogues.
Low stringency alignments with the TIGR plant gene indices
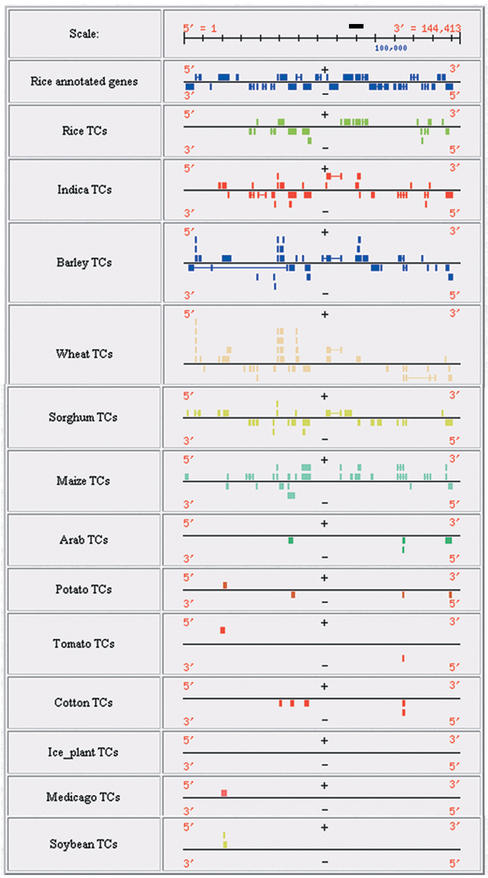
With the exception of rice and Arabidopsis thaliana, genome sequence data for other plant species is primarily comprised of EST data. These EST sequences have been condensed into gene indices that represent clustered assemblies of the expressed portion of the genome (7; http://www.tigr.org/tdb/tgi/) and provide a robust resource to sample these plant genomes. To provide a comprehensive view of sequence similarity among plant species, we have aligned the publicly available rice BAC/PAC sequences with 13 TIGR plant gene indices that represent 12 species of monocotyledonous and dicotyledonous plants (Fig. 4; http://www.tigr.org/tdb/tgi/ogi/alignTC.html). Included in these displays are alignments with two rice gene indices; one from the rice ESTs available in GenBank and one index build exclusively of ESTs from the indica subspecies of rice available from the Beijing Genomics Institute (BGI; http://btn.genomics.org.cn/rice). The inclusion of the indica ESTs allows for a direct comparison of EST representation in the primarily japonica-derived public ESTs in GenBank with the indica-derived ESTs available from the BGI. Along with the alignments with the gene indices, the models generated from our automated annotation of the BAC/PACs are displayed allowing for comparison between automated annotation results with alignments with expressed sequences. In addition to the graphical displays provided through these web pages, the alignments of the BAC/PACs with the gene indices are available through our rice DAS (Fig. 3; http://www.tigr.org/tdb/e2k1/osa1/irgsp/DAS.shtml).
Figure 4.
Alignment of rice BAC/PACs with the TIGR plant gene indices. Alignment of OSJNBa0015N08 with 12 TIGR plant gene indices and a gene index generated from the BGI indica EST sequences are shown along with the gene models generated for this BAC from our automated annotation pipeline. This is the same BAC as in Figure 2 and the position of working model #19 is indicated by a solid black bar.
Inclusion of rice in the TIGR Eukaryotic Gene Orthologue Database
Using the reciprocal top hit method (16), we have created a eukaryotic gene ortholog database that contains putative orthologues and paralogues among the 53 species represented in the TIGR gene indices (http://www.tigr.org/tdb/tgi/ego/index.shtml). In the current build of the EGO (1.0), there are a total of 6729 unique members with the most frequent orthologue pairs being rice–maize (5437 pairs) and rice–Arabidopsis (4789 pairs).
Syntenic alignments between rice and maize
It has been well documented that rice is highly syntenic with other cereal species such as maize, wheat, barley, sugarcane, sorghum, and millet (2). These studies were performed using a limited number of conserved orthologous markers and provide a low resolution syntenic map between these cereal species. With access to the complete rice genome, it is possible to increase the resolution of the syntenic map between rice and these other agronomically significant crop species. To align the rice genome with cereal sequences, we first anchored 2464 rice BAC/PAC sequences to the rice genetic map using in silico alignments with 13 251 available rice genetic markers. We then searched these rice BAC/PACs along with the rice genetic markers against 1259 anchored maize markers available from MaizeDB (http://www.agron.missouri.edu). Using a high stringency cutoff criteria, we aligned 350 maize markers with rice sequences (markers or BAC/PACs). In total, ∼53% of the alignments were on the syntenic maize chromosome. The data from these alignments can be accessed through a web-based interface (http://www.tigr.org/tdb/e2k1/osa1/maize/description.shtml) and provide a starting point for comparative mapping between these two significant cereal species.
CONCLUSIONS
In the TIGR Rice Genome Annotation Resource, we have annotation data for 369 Mb of rice genomic DNA representing ∼77% of the rice genome. Upon the completion of the draft phase of the IRGSP effort in December 2002 (http://rgp.dna.affrc.go.jp/rgp/press_releas20011225.htm), we will add the remainder of the BAC/PACs to our annotation pipeline, thereby, providing a more complete resource for plant biologist. Through identification of related sequences in other plant species and alignments of the rice genome with syntenic markers, we have provided the foundation to leverage the rice genome sequence to other plant species. We have provided access to these data through web-based interfaces, FTP download of flatfiles, and a DAS server thereby providing a rich resource for data-mining the publicly available rice genome sequence.
Acknowledgments
ACKNOWLEDGEMENTS
Funding for the work was provided by a grant to C.R.B. from the US Department of Agriculture (99-35317-8275), the National Science Foundation (DBI998282), and the US Department of Energy (DE-FG02-99ER20357). The authors wish to thank Lowell Umayan, Jeremy Peterson, Qi Yang, Brian Haas, Sam Angiouli, Owen White, Michael Heaney, Susan Lo, Vadim Sapiro, Billy Lee, Jeff Shao, Corey Irwin, Rajeev Kramchedu, Jacqueline Neubrech, Mark Sengamalay and Eddy Arnold for their bioinformatic and IT support.
REFERENCES
- 1.Maclean J. (1997) Rice Almanac. International Rice Research Institute, Manilla Philippines.
- 2.Gale M.D. and Devos,K.M. (1998) Comparative genetics in the grasses. Proc. Natl Acad. Sci. USA, 95, 1971–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barry G.F. (2001) The use of the Monsanto draft rice genome sequence in research. Plant Physiol., 125, 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sasaki T. and Burr,B. (2000) International Rice Genome Sequencing Project: the effort to completely sequence the rice genome. Curr. Opin. Plant Biol., 3, 138–141. [DOI] [PubMed] [Google Scholar]
- 5.Goff S.A., Ricke,D., Lan,T.H., Presting,G., Wang,R., Dunn,M., Glazebrook,J., Sessions,A., Oeller,P., Varma,H. et al. (2002) A draft sequence of the rice genome (Oryza sativa L ssp. japonica) Science, 296, 92–100. [DOI] [PubMed] [Google Scholar]
- 6.Yu J., Hu,S., Wang,J., Wong,G.K., Li,S., Deng,Y., Dai,L., Zhou,Y., Zhang,X. et al. (2002) A draft sequence of the rice genome (Oryza sativa L ssp. indica). Science, 296, 79–92. [DOI] [PubMed] [Google Scholar]
- 7.Quackenbush J., Liang,F., Holt,I., Pertea,G. and Upton,J. (2000) The TIGR gene indices: reconstruction and representation of expressed gene sequences. Nucleic Acids Res., 28, 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowe T.M. and Eddy,S.R. (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res., 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dowell R.D., Jokerst,R.M., Day,A., Eddy,S.R. and Stein,L. (2001) The Distributed Annotation System. BMC Bioinformatics, 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arumuganathan K. and Earle,E.D. (1991) Nuclear DNA content of some important plant species. Plant Mol. Biol. Reporter, 9, 208–218. [Google Scholar]
- 11.Nielsen H., Engelbrecht,J., Brunak,S. and von Heijne,G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng., 10, 1–6. [DOI] [PubMed] [Google Scholar]
- 12.Krogh A., Larsson,B., von Heijne,G. and Sonnhammer,E.L.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol., 305, 567–580. [DOI] [PubMed] [Google Scholar]
- 13.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falquet L., Pagni,M., Bucher,P., Hulo,N., Sigrist,C.J., Hofmann,K. and Bairoch,A. (2002) The PROSITE database, its status in 2002. Nucleic Acids Res., 30, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goff S.A. (1999) Rice as a model for cereal genomics. Curr. Opin. Plant Biol., 2, 86–89. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y., Sultana,R., Pertea,G., Cho,J., Karamycheva,S., Tsai,J., Parvizi,B., Cheung,F., Antonescu,V., White,J., Holt,I., Liang,F. and Quackenbush,J. (2002) Cross-referencing eukaryotic genomes: TIGR Orthologous Gene Alignments (TOGA). Genome Res., 12, 493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]