Abstract
The CATH database of protein domain structures (http://www.biochem.ucl.ac.uk/bsm/cath_new) currently contains 34 287 domain structures classified into 1383 superfamilies and 3285 sequence families. Each structural family is expanded with domain sequence relatives recruited from GenBank using a variety of efficient sequence search protocols and reliable thresholds. This extended resource, known as the CATH-protein family database (CATH-PFDB) contains a total of 310 000 domain sequences classified into 26 812 sequence families. New sequence search protocols have been designed, based on these intermediate sequence libraries, to allow more regular updating of the classification.
Further developments include the adaptation of a recently developed method for rapid structure comparison, based on secondary structure matching, for domain boundary assignment. The philosophy behind CATHEDRAL is the recognition of recurrent folds already classified in CATH. Benchmarking of CATHEDRAL, using manually validated domain assignments, demonstrated that 43% of domains boundaries could be completely automatically assigned. This is an improvement on a previous consensus approach for which only 10–20% of domains could be reliably processed in a completely automated fashion. Since domain boundary assignment is a significant bottleneck in the classification of new structures, CATHEDRAL will also help to increase the frequency of CATH updates.
DESCRIPTION OF THE CATH HIERARCHY AND CURRENT POPULATION STATISTICS
The CATH database is a hierarchical classification of domains into sequence and structure based families and fold groups. Table 1 shows the population of the latest release of CATH (Version 2.4). In the lowest level of the hierarchy, sequences are clustered according to significant sequence similarity (>35% identity and above, the S-Level). At higher levels, domains are grouped according to whether they share significant sequence, structural and/or functional similarity (homologous superfamilies, H-Level) or just structural similarity (fold or topology group, the T-level). Fold groups sharing similar architectures, that is similarities in the arrangements of their secondary structures regardless of connectivity are then merged into the common architectures (the A-Level). At the top of the hierarchy, domains are clustered depending on their class, that is the percentage of α-helices or β-strands (the C-Level).
Table 1. Populations of the different levels in the CATH hierarchy.
| Class | 1 | 2 | 3 | 4 | 5 | Total |
|---|---|---|---|---|---|---|
| A | 5 | 19 | 13 | 1 | n/a | 38 |
| T | 219 | 133 | 346 | 77 | n/a | 775 |
| H | 381 | 270 | 653 | 82 | n/a | 1386 |
| S | 770 | 768 | 1647 | 100 | 850 | 3285 |
| All | 7153 | 10 112 | 16 279 | 743 | 7881 | 34 287 |
IMPROVED CLASSIFICATION PROTOCOLS
Below we describe some new protocols which increase the speed of classifying newly determined protein structures in the CATH database. These include a new method for rapid detection of homologues by intermediate sequence searching techniques, automatic method for domain boundaries in multidomain proteins and a new protocol for homologue detection.
IMPROVED CLASSIFICATION PROTOCOLS BASED ON INTERMEDIATE SEQUENCE SEARCHING
Profile based methods for sequence comparison were developed in the early 80s and allowed recognition of more distant homologues, than pairwise based approaches [3]. Benchmarking of several publicly available methods, including those using position specific score matrices and Hidden Markov models have been undertaken by several groups (4,5). These approaches used datasets of distant homologues selected from the structural classifications, such as SCOP and CATH, to determine the sensitivity of various profile based methods e.g. PSI-BLAST, (6). Up to 20% more remote homologues could be detected (from ∼30% for pairwise methods up to ∼55% for profiles), depending on the dataset and search method used.
We have benchmarked several powerful profile based search methods [PSI-BLAST, IMPALA (6) and SAM-T99 (7,8)] and optimized the parameters for reliable detection of distant homologues with a low error rate (<0.1%). The DomainFinder protocol uses these approaches to match CATH domain sequences to relatives in the non-redundant GenBank database [DomainFinder, (9,10)] thereby currently recruiting nearly 310 000 additional sequence domains into the CATH database. Pairwise sequence comparison, using standard dynamic programming approaches [HOMOL, (11)], followed by single linkage clustering, currently clusters these sequences into 26 812 sequence families in the database (S-Levels). The extended CATH database is referred to as the CATH-protein family database (CATH-PFDB).
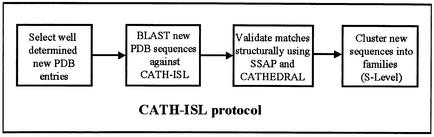
The establishment of the CATH-PFDB has enabled a novel sequence search protocol, based on intermediate sequence searching (Fig. 1) to be adopted as the first stage of homologue recognition in the classification of newly determined structures in the CATH database. In this procedure, simple pairwise alignment methods such as BLAST can be used to scan query sequences against an intermediate sequence library of non-identical sequences from the CATH-PFDB (CATH-ISL). Benchmarking, using a stringent data-set of remote structural homologues from CATH, has shown that intermediate sequence searching approaches based on ‘BLASTing’ the CATH-ISL perform as well as profile based approaches, like PSI-BLAST or IMPALA (9) but are considerably faster. In the CATH intermediate sequence search protocol (Fig. 1), new structures matching CATH domains in the CATH-ISL are identified and then validated by structure comparison. Once recruited to a particular homologous superfamily, sequences are clustered into sequence families.
Figure 1.
Flowchart of the new CATH protocol which uses intermediate sequence searching to classify newly determined structures. Sequences are BLASTed against an intermediate sequence library (CATH-PFDB) and potential matches are structurally validated before clustering into sequence families.
Currently, up to 70% of newly determined protein structures can be classified using these sequence based approaches, considerably reducing the database scans which must be performed using the computationally more expensive structure alignment methods.
AN AUTOMATED METHOD FOR DOMAIN BOUNDARY ASSIGNMENT BASED ON FOLD RECURRENCE
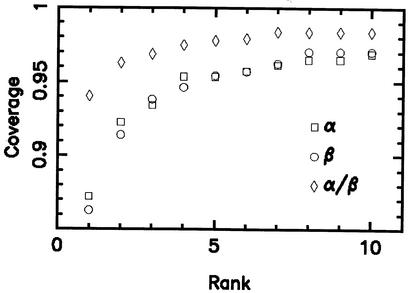
To detect very remote homologues unrecognized by sequence based approaches, a rapid method of structure comparison [GRATH, (1)] is used as a pre-filter to a slower but more accurate method [SSAP, (12)]. GRATH was inspired by the graph theoretical approaches developed by Artymiuk andco-workers in the early 1990s (13). Graph theory is used to compare secondary structure vectors between proteins and a robust statistical framework allows reliable assessment of the significance of any structural match observed (14). Furthermore, benchmarking of a completely automated protocol employing GRATH showed that for 98% of the structures in a large test dataset the correct fold group was correctly identified within the top 10 matches returned from a database scan (Fig. 2).
Figure 2.
Coverage plots showing the proportion of structures which are assigned to the correct fold group, using the GRATH algorithm, within the top N ranked matches returned from a database search of non-identical representatives from CATH.
GRATH is, therefore, used as a front end filter for the residue based structure comparison method SSAP (12) which has traditionally been used for classifying structures in CATH and which returns an accurate residue based alignment for structures belonging to the same fold group. This considerably increases the speed of classifying newly determined structures in CATH because GRATH is typically up to 1000 times faster than SSAP.
The statistical framework developed for GRATH has enabled another new CATH protocol for automatic recognition of domain boundaries in those multidomain proteins comprising domains with folds already classified in CATH. The use of fold recurrence in domain boundary recognition has already been successfully exploited by the PUU algorithm (15) for classifying structures in the DALI Domain Database (16) and also in the SCOP classification, where manual assignment is performed.
We have developed the CATHEDRAL algorithm which exploits GRATH and the statistical framework associated with GRATH, to iteratively recognize and extract the most significant fold matches within a multidomain protein. Regions matching previously classified structural domains in CATH are accurately aligned using the SSAP algorithm to further validate the match and then ‘removed’ from the multidomain structure. The remaining structure is then rescanned against the CATH fold library until all matching domains have been identified. For 70% of cases in a large test dataset, domain boundaries are accurately assigned with an accuracy of +/− 10 residues. For the remaining 30% of cases, the fold adopted by the core of the domain was correctly identified. However, because these matches often involved very remote homologues (<20% sequence identity), domain embellishment had frequently occurred (17), resulting in some cases in significant increases in the size of the domain. Variation across fold groups can be even more significant. In these cases, manual validation and adjustment must be performed to refine the boundaries.
Use of stringent thresholds on E-value and overlap criteria, together with the constraint that all the domains in a multidomain structure must be assigned folds, allows completely automated domain boundary assignment for, on average, 40–50% of newly determined multidomain structures, classified in CATH. The previous protocol for domain boundary assignment in CATH [DBS, (3)] sought a consensus between three completely independent automatic methods of domain boundary recognition [PUU, DOMAK, DETECTIVE, see (18) and references therein]. Though each individual method cited up to 70–80% accuracy, the methods rarely agreed over a significant portion of the domain so that application of DBS typically only allowed completely automatic boundary assignment for between 10–20% of multidomain proteins.
Since structures are much more highly conserved than sequence during evolution, it is generally easier and more reliable to assign boundaries using structural data and information on boundary domains is, therefore, one of the most important types of derived data that the structural classifications provide. The improvement in the percentage of domains automatically assigned using CATHEDRAL will significantly increase the speed of classification in CATH as domain boundary assignment is one of the major bottlenecks.
There are 775 folds within the CATH database and currently 80% of domains within multidomain structures in CATH possess folds which recur as single domains or in different multidomain contexts. This proportion is likely to increase as the number of known structures increases and as structural genomics initiatives selectively target putative novel folds for structure determination. Therefore, the proportion of domain boundaries which can be automatically assigned by CATHEDRAL will also increase as the structural genomics initiatives proceed.
EXPANSION OF FUNCTIONAL ANNOTATIONS IN CATH-PFDB AND NEW PROTOCOLS FOR HOMOLOGUE DETECTION
Functional annotations for each family and superfamily in the CATH-PFDB have been extended by recruiting relevant functional data from the PDB, SWISS-PROT, GenProtEC, GenBank, Enzyme (EC) and GO databases. The nearly tenfold expansion in the CATH-PFDB database (from 34 000 CATH structural domain sequences to ∼310 000 CATH-PFDB, including related GenBank sequences) has significantly increased the amount of functional data available for a particular family or superfamily. This has enabled extensive analysis of the evolution of function in protein superfamilies (19).
A new text-mining method has been developed (Bennett, personal communication) for comparing the functional annotations of newly determined structures with those of putative relatives from the family and superfamily into which the query structure has been classified. Statistics of the frequencies of particular functional keywords and classification numbers are first compiled for each family and then the similarity of keywords belonging to the query structure are assessed and scored depending on the frequency of the matched word. Currently 50% of remote homologues (<35% sequence identity or E-value>0.01 by PSI-BLAST match), which have been assigned to a particular superfamily by structure matching, can be validated using this approach. The increase in functional annotations in the CATH-PFDB contributes significantly to the success of this approach.
Sequence, structure and functional data from the CATH database are stored in an ORACLE 9i relational database (20), together with genome identifiers and information of gene taxonomy. The use of ORACLE allows metalevels to be constructed capturing the relationships existing between different levels in the hierarchical classification e.g. fold group and superfamily, domain and sequence segment. It also enables the design of more sophisticated user query interfaces for selecting information useful for functional genomics. For example, all the structural and functional annotations associated with genes selected from a specific genome or genes being co-expressed in a transcriptomics experiment.
Acknowledgments
ACKNOWLEDGEMENTS
F.M.G.P., A.P.H., D.L., I.S. and C.A.O. all acknowledge the Medical Research Council for their funding. J.E.B. is currently supported by funding from the NIH. A.S. acknowledges supports from the Biotechnology and Biological Research Council and C.F.B. acknowledges support from the Wellcome Trust for research described in this manuscript.
REFERENCES
- 1.Harrison A., Pearl,F., Sillitoe,I., Slidel,T., Mott,R., Thornton,J. and Orengo,C. (2002) A fast method for reliably recognising the fold of a protein structure. Submitted to Bioinformatics. [DOI] [PubMed]
- 2.Harrison A., Pearl,F., Sillitoe,I., Thornton,J. and Orengo,C. (2002) CATHEDRAL: an effective algorithm to delineate previously seen folds within a multi-domain structure. In preparation.
- 3.Jones D.T., Taylor,W.R. and Thornton,J.M. (1992) A new approach to protein fold recognition. Nature, 358, 86–89. [DOI] [PubMed] [Google Scholar]
- 4.Brenner S.E., Chothia,C. and Hubbard,T.J.P. (1998) Assessing sequence comparison methods with reliable structurally identified distant evolutionary relationships. Proc. Natl Acad. Sci. USA, 95, 6073–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pearl F., Todd,A.E., Bray,J.E., Martin,A.C., Salamov,A.A., Suwa,M., Swindells,M.B., Thornton,J.M. and Orengo,C.A. (2000) Using the CATH domain database to assign structures and functions to the genome sequences. Biochem. Soc. Trans., 28, 269–275. [DOI] [PubMed] [Google Scholar]
- 6.Altschul S.F., Bundschuh,R., Olsen,R. and Hwa,T. (2001) The estimation of statistical parameters for local alignment score distributions. Nucleic Acids Res., 29, 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karplus K., Sjolander,K., Barrett,C., Cline,C., Hausser,D., Hughey,R., Holm,L. and Sander,C. (1997) Predicting protein structure using hidden Markov models. Proteins, 1(Suppl), 134–139. [DOI] [PubMed] [Google Scholar]
- 8.Karplus K., Barrett,C. and Hughey,R. (1998) Hidden Markov models for detecting remote protein homologies. Bioinformatics, 14, 846–856. [DOI] [PubMed] [Google Scholar]
- 9.Pearl F.M., Martin,N., Bray,J., Buchan,D.W.A., Harrison,A.P., Lee,D., Reeves,G.A., Shepherd,A.J., Sillitoe,I., Todd,A.E., Thornton,J.M. and Orengo,C.A. (2001) A rapid classification protocol for the CATH Domain Database to support structural genomics. Nucleic Acids Res., 29, 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearl F.M., Lee,D., Bray,J.E., Buchan,D.W., Shepherd,A.J. and Orengo,C.A. (2002) The CATH extended protein-family database: providing structural annotations for genome sequences. Protein Sci., 11, 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orengo C., Michie,A., Jones,S., Jones,D., Swindells,M. and Thornton,J. (1997) CATH—a hierarchic classification of protein domain structures. Structure, 5, 1093–1108. [DOI] [PubMed] [Google Scholar]
- 12.Taylor W.R. and Orengo,C.A. (1989) Protein structure alignment. J. Mol. Biol., 5, 1–22. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell E.M., Artymiuk,P.J., Rice,D.W. and Willett,P. (1990) Use of techniques derived from graph theory to compare secondary structure motifs in proteins. J. Mol. Biol., 212, 151–166. [DOI] [PubMed] [Google Scholar]
- 14.Harrison A., Pearl,F., Mott,R., Thornton,J. and Orengo,C. (2002) Quantifying the similarities within fold space. J. Mol. Biol., 323, 909–926. [DOI] [PubMed] [Google Scholar]
- 15.Holm L. and Sander,C. (1994) Parser for protein folding units. Proteins, 19, 256–268. [DOI] [PubMed] [Google Scholar]
- 16.Holm L. and Sander,C. (1999) Protein folds and families: sequence and structure alignments. Nucleic Acids Res., 27, 244–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orengo C.A., Sillitoe,I., Reeves,G. and Pearl,F.M. (2001) Review: what can structural classifications reveal about protein evolution? J. Struct. Biol., 134, 145–165. [DOI] [PubMed] [Google Scholar]
- 18.Jones S., Stewart,M., Michie,A., Swindells,M.B., Orengo,C. and Thornton,J.M. (1998) Domain assignment for protein structures using a consensus approach: characterization and analysis. Protein Sci., 7, 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Todd A., Orengo,C. and Thornton,J. (2001) Domain assignment for protein structures using a consensus approach: characterization and analysis. J. Mol. Biol., 307, 1113–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shepherd A.L., Martin,N., Johnson,R.G., Kellam,P. and Orengo,C.A. (2002) PFDB: A generic protein family database integrating the CATH domain structure database with sequence based protein family resources. Bioinformatics, in press. [DOI] [PubMed] [Google Scholar]