Abstract
UV vision has profound effects on the evolution of organisms by affecting such behaviors as mating preference and foraging strategies. Despite its importance, the molecular basis of UV vision is not known. Here, we have transformed the zebra finch UV pigment into a violet pigment by incorporating one amino acid change, C84S. By incorporating the reverse mutations, we have also constructed UV pigments from the orthologous violet pigments of the pigeon and chicken. These results and comparative amino acid sequence analyses of the pigments in vertebrates demonstrate that many avian species have achieved their UV vision by S84C.
UV vision has been found in many fish, amphibian, avian, reptilian, and mammalian species (1). The petals of bird-pollinated flowers have substantial UV reflectance, which provide attractive targets to birds with UV vision (2). Similarly, the scales of fish and the feathers of birds often reflect UV, improving the visibility of their body coloration patterns (3, 4). Indeed, UV vision has been used in a variety of situations, including social signaling (5), hunting (6), nectar localization (2), and mate-choice (7–9). UV vision is determined solely by visual pigments (10–12), each of which consists of the chromophore, 11-cis-retinal, and a transmembrane protein, opsin. The retinal visual pigments in vertebrates are classified into five evolutionarily distinct groups: (i) rhodopsin (RH1), (ii) RH1-like (RH2), (iii) short wavelength-sensitive (SWS1), (iv) SWS1-like (SWS2), and (v) long wavelength-sensitive (LWS) or middle wavelength-sensitive (MWS) (LWS/MWS) groups (13–15). The phylogenetic relationship of these pigments is given by ((((RH1, RH2), SWS2), SWS1), LWS/MWS), and the UV pigments that absorb light maximally (λmax) at around 360 nm belong to the SWS1 group (15, 16).
So far, the UV opsin genes of mouse, rat, chameleon, and goldfish have been characterized. By comparing the amino acid sequences deduced from the nucleotide sequences of these and related opsin genes, several amino acids have been suggested to be important in achieving UV-sensitivity (12, 17, 18). However, the molecular bases of UV vision are still unknown. This is because it has been difficult to regenerate UV pigments in vitro, and, consequently, mutagenesis experiments could not be conducted. Indeed, it was only a few years ago that the UV pigments have been regenerated successfully (18–20).
Here, we report the cloning of the UV opsin cDNA and regeneration of the UV pigment of the zebra finch (Taeniopygia guttata). We then describe the processes of the transformation of the zebra finch UV pigment into a violet pigment and the reverse changes of the violet pigments of chicken and pigeon. These experiments and amino acid sequence analyses of the SWS1 pigments in vertebrates show that only one amino acid change was responsible for the development of UV pigments in birds.
Materials and Methods
Cloning and Sequencing of cDNA clones.
A cDNA library was constructed in the λZAPII vector by using 5 μg of poly(A) mRNA from 20 zebra finch retinas and Poly(A) Quick mRNA isolation kit, ZAP-cDNA Synthesis kit, and Gigapack III Gold extracts, following the protocols supplied by the manufacturer (Stratagene). In the initial screening, about 4 × 105 recombinant plaques were transferred to nylon membrane (Hybond-N+, Amersham) for hybridization with a mixture of 32P-random-labeled human blue opsin cDNA (hs37, a gift from J. Nathans, Johns Hopkins Univ., Baltimore). Hybridization was carried out at 55°C, and hybridized membranes were washed four times (30 min each) in 1× SSC (0.15 M NaCl/0.015 M Na3 citrate)/0.1% SDS at 55°C. Of the 16 cDNA clones isolated, one clone was found to contain the entire coding region. This clone was subcloned into pBluescript SK(−) vector and was sequenced by cycle sequencing reactions using the Sequitherm Excel II Long-Read kits (Epicentre Technologies, Madison, WI) with dye-labeled M13 forward and reverse primers. Reactions were run on a Li-Cor 4200LD automated DNA sequencer (Li-Cor, Lincoln, NE).
Regeneration of Visual Pigments, Site-Directed Mutagenesis, and Spectral Analyses.
In addition to that of the zebra finch, total RNA was also isolated from the retinas of a chicken. To conduct reverse transcription–PCR, two sets of primers were prepared: 5′-GGAATGAATTCCACCATGGACGAGGAAGAG-3′ (Forward) and 5′-GCAGAGGTCGACCTGGGGCGGACCTGGCTG-3′ (Reverse) for the zebra finch UV pigment and 5′-ATTATTGAATTCCACCATGTCATCGGACGACGA-3′ (F) and 5′-TATATAGTCGACGGACCAACTTGGCTGGAGGACACGGA-3′ (R) for the chicken violet pigment. The first strand cDNA synthesis was carried out at 42°C for 1 h in a total volume of 20 μl containing reaction buffer (10 mM Tris⋅HCl, pH 9.0/1 mM MgCl2/50 mM KCl/0.1% Triton X-100), 1 mM dNTPs, 5 μM reverse primers, 20 units of RNasin (Promega), and 200 units of SuperScript II Reverse transcriptase (GIBCO/BRL). The resulting cDNA was combined with the same reaction buffer containing 200 mM dNTPs, 1 μM each forward and reverse primers, and 5 units of Taq polymerase (Promega) in a total volume of 100 μl. PCR amplification was performed by 30 cycles at 92°C for 45 sec, 55°C for 60 sec, and 72°C for 90 sec. At each cycle, the duration of the extension reaction was progressively extended by 3 sec. After the final extension step at 72°C for 10 min, the PCR products were resolved in 1.5% agarose gel electrophoresis. The amplified cDNA clones were sequenced to rule out spurious mutations and were subcloned into the expression vector pMT5. This plasmid was expressed in COS1 cells by transient transfection. The pigment was regenerated by incubation with 11-cis-retinal and was purified in buffer consisting of 50 mM N-(2-hydroxyethyl) piperazine-N′-2-ethanesulfonic acid (pH 6.6), 140 mM NaCl, 3 mM MgCl2, 20% (wt/vol) glycerol, and 0.1% dodecyl maltoside (21).
Mutants were generated by using QuickChange site-directed mutagenesis kit (Stratagene). All DNA fragments that were subjected to mutagenesis were sequenced to rule out spurious mutations. UV visible absorption spectra of visual pigments were recorded at 20°C, using a Hitachi (Tokyo) U-3000 dual beam spectrophotometer. Visual pigments were bleached by a 366-nm UV light illuminator. Recorded spectra were analyzed by using sigmaplot software (Jandel, San Rafael, CA).
Sequence Data Analysis.
At present, 15 SWS1 pigments have been characterized for their amino acid sequences as well as absorption spectra (Table 1). By using more evolutionarily distantly related RH1, RH2, SWS2, and LWS/MWS pigments (Table 1) as the outgroup, the rooted phylogenetic tree for the SWS1 pigments in vertebrates was constructed. The number (K) of amino acid substitutions per site was estimated from K = −ln (1 − p), where p is the proportion of different amino acids for a pair of sequences. The phylogenetic tree was reconstructed by applying the neighbor-joining method (22) to the K values. The reliability of the phylogenetic tree was evaluated by the bootstrap analysis with 1,000 replications (23). In addition, by using the corresponding nucleotide sequences, the numbers of nucleotide substitutions per site were estimated for all pairs (24), from which the phylogenetic tree of the SWS1 pigments was also constructed by using the NJ method.
Table 1.
SWS1 and paralogous visual pigments in vertebrates
| Group | Visual pigment | GenBank accession no. | Group | Visual pigment | GenBank accession no. |
|---|---|---|---|---|---|
| SWS1 | Goldfish (P359) | D85863 | SWS1 | Marmoset (P424) | L22218 |
| Zebrafish (P362) | AF109373 | Mouse (P359) | U49720 | ||
| Clawed frog (P425) | U23463 | Rat (P358) | U63972 | ||
| Chameleon (P358) | AF134192 | RH1 | Bovine (P500) | M21606 | |
| Pigeon (P393) | AF149234 | RH2 | Goldfish (P511) | L11865 | |
| Zebra finch (P359) | AF222331 | SWS2 | Goldfish (P441) | L11864 | |
| Canary (P366) | Ref. 12 | Chameleon (P437) | AF133907 | ||
| Chicken (P415) | M92039 | LWS/MWS | Goldfish (P559) | L11867 | |
| Parakeet (P371) | Y11787 | Clawed frog (P611) | U90895 | ||
| Human (P413) | M13295 | Chameleon (P561) | U08131 | ||
| Squirrel monkey (P433) | U53875 | Pigeon (P558) | AF149243 | ||
| Macaque (P415) | AF158977 | Human (P560) | M1330 |
The number after P in the parenthesis refers to λmax. Bovine, Bos taurus; Canary, Serinus canaria; Chameleon, Anolis carolinensis; Chicken, Gallus gallus; Clawed frog, Xenopus laevis; Goldfish, Carassius auratus; Human, Homo sapiens; Macaque, Macaca fascicularis; Marmoset, Callithrix jacchus; Mouse, Mus musculus; Parakeet, Melopsittacus undulatus; Pigeon, Columba livia; Rat, Rattus norvegicus; Squirrel monkey, Saimiri boliviensis; Zebra finch, Taeniopygia guttata; Zebrafish, Danio rerio.
Results and Discussion
Absorption Spectrum of the Zebra Finch UV Pigment.
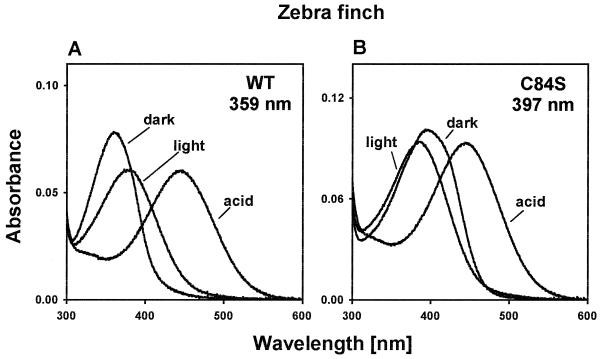
Screening a λZAP II zebra finch retinal cDNA library with human blue opsin cDNA as the probe, we have obtained one complete clone. The opsin deduced from this cDNA sequence consists of 346 amino acids (GenBank accession no. AF222331). By expressing the opsin in cultured cells and reconstituting the product with 11-cis-retinal, we have regenerated the zebra finch pigment. This pigment is sensitive to wavelengths between 320 and 400 nm, with a maximum value (λmax) at 359 ± 1 nm (Fig. 1A), which is consistent with a previous estimate, 360–380 nm, for the zebra finch UV pigment determined by microspectrophotometry (11).
Figure 1.
Absorption spectra of the zebra finch pigments. (A) Absorption spectrum for the wild-type pigment (WT) and those after exposure to light (light) and H2SO4 (acid). (B) Absorption spectrum for the mutant pigment (C84S) and those after exposure to light and H2SO4. The ratios of the protein absorption peak (not shown) to the pigment absorption peak were 2.56 and 3.07 for the wild-type and mutant pigments, respectively.
When the regenerated wild-type pigment was exposed to UV light, a new peak absorption at ≈380 nm was achieved (Fig. 1A). This means that 11-cis-retinal in the pigment was isomerized by light and all-trans-retinal was released. Furthermore, when the pigment was denatured by sulfuric acid (H2SO4) at pH 1.8 in the dark, the resulting spectrum had the peak absorption at ≈440 nm (Fig. 1A), typical of a protonated Schiff base 11-cis-retinal free in solution. Because acid has no effect on the absorbance of free 11-cis-retinal, this result shows that the observed peak at 359 nm is generated by opsin covalently linked to 11-cis-retinal in a Schiff base linkage (18–20). Thus, these control experiments demonstrate that we have indeed regenerated the UV pigment of the zebra finch.
Phylogenetic Relationship of the SWS1 Pigments in Vertebrates.
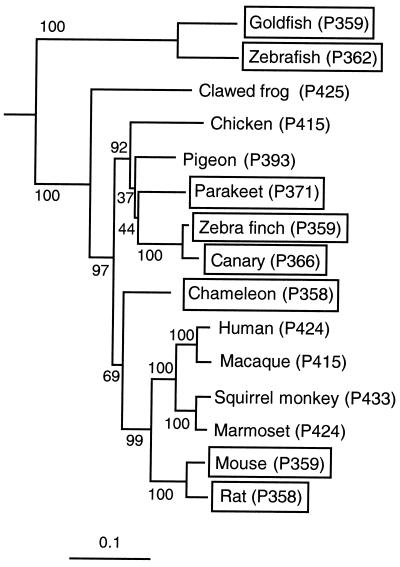
Phylogenetic analyses show that the UV pigment of the zebra finch, referred to as zebra finch (P359), belongs to the SWS1 group (Fig. 2). The topology of the phylogenetic tree of the SWS1 pigments is mostly consistent with that of the organismal tree, where mammals are more closely related to reptiles/birds, amphibians, and fishes, in that order. The phylogenetic positions of most of the SWS1 pigments have high levels of bootstrap support and are highly reproducible (Fig. 2). Note, however, that chameleon (P358) pigment is more closely related to the mammalian pigments than to the avian pigments. The bootstrap support for the clustering of the chameleon and mammalian pigments is only 69%, and the exact phylogenetic position of chameleon (P358) pigment cannot be established. Thus, considering the relationships of organisms (25), it seems reasonable to assume that chameleon (P358) pigment is more closely related to the avian pigments than to the mammalian pigments. The clustering of chicken (P415), pigeon (P393), parakeet (P371), zebra finch (P359), and canary (P366) pigments is well supported by the bootstrap analysis, but the exact phylogenetic positions of these pigments cannot be established because of poor bootstrap supports (Fig. 2). Nevertheless, it is interesting to observe that the three avian UV pigments cluster as one group, suggesting the possibility that the evolution of the avian UV pigments occurred only once in the avian lineage.
Figure 2.
The phylogenetic tree for vertebrate SWS1 pigments. The numbers after P refer to λmax, and those next to the different branches are clustering percentage supports generated by 1,000 bootstrap replicates. UV pigments are boxed. The horizontal bar at the bottom indicates evolutionary distance measured by the number of amino acid replacements per site.
The phylogenetic tree of the SWS1 pigments can also be constructed by considering the corresponding nucleotide sequences. Unfortunately, the nucleotide sequence of the canary (P366) opsin gene is not available, and, therefore, it has to be excluded from the analysis. The tree topology obtained by applying the neighbor-joining method to the nucleotide sequences of the remaining opsin genes differs from that of Fig. 2 at two points. First, as suspected, chameleon (P358) pigment now clusters with the avian pigments with the bootstrap support of 99% (results not shown). Second, the “nucleotide tree” suggests the phylogenetic relationship (((zebra finch (P359), pigeon (P393)), parakeet (P371)), chicken (P415)), and the phylogenetic positions of the parakeet and pigeon pigments are now exchanged. The cluster of the avian pigments is highly reproducible with a bootstrap support of 92%, but their exact phylogenetic positions still cannot be resolved.
Molecular Evolution of the Avian UV Pigments: Hypothesis.
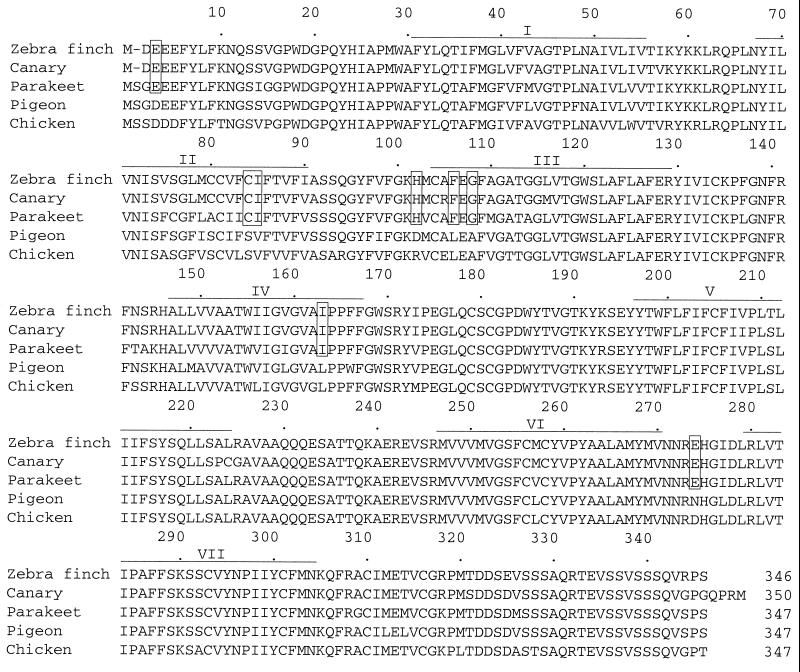
The absorption spectra of visual pigments are determined by the interactions between 11-cis-retinal and opsin. Thus, to elucidate the evolutionary divergence of UV pigments and violet pigments, we need to identify the amino acids that may be responsible for the functional differentiation of the two types of pigments. When the amino acids of the 15 SWS1 pigments are compared site by site, we cannot find any amino acid that is common only to the UV pigments, suggesting that the λmax values of the UV pigments in various vertebrates have been achieved by different mechanisms. Under this circumstance, the functional divergence of UV and violet pigments can be studied most effectively by comparing the two types of pigments in a particular lineage of organisms. At present, the UV and violet pigments can be compared only in two groups: birds and mammals (Fig. 2). When the two types of pigments in birds are compared, 8 amino acids are found to be conserved among parakeet (P371), zebra finch (P359), and canary (P366) pigments (Fig. 3) whereas when those in mammals are compared, 24 amino acids are conserved between the mouse and rat UV pigments. Clearly, it is much simpler to study the molecular basis of the UV pigments in birds than those in mammals.
Figure 3.
Alignment of the amino acid sequences of the SWS1 pigments of birds. Zebra finch, canary, parakeet, pigeon, and chicken indicate zebra finch (P359), canary (P366), parakeet (P371), pigeon (P393), and chicken (P415) pigments, respectively. Gaps necessary to increase the sequence similarity are indicated by dashes (-). Our analysis of the chicken opsin cDNA shows that the nucleotide sequences at codon position 113 are ACC (encoding threonine) instead of CAC (histidine) reported in the GenBank database (M92039). Seven putative transmembrane regions (40) are indicated by horizontal lines, and the avian UV pigment-specific amino acids are boxed.
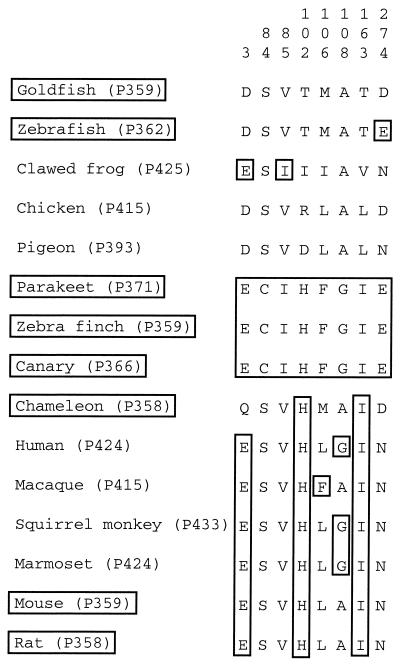
When the eight amino acids conserved among the three avian UV pigments are compared with those at the corresponding sites of the other SWS1 pigments, the UV pigment-specific amino acids E3, I85, H102, F106, G108, and I163 are shared by violet pigments (Fig. 4) whereas E274 is located in the nontransmembrane region and is unlikely to interact with the chromophore (Fig. 3). However, C84 is associated distinctly with UV pigments and cannot be found in other SWS1 pigments. This strongly suggests that C84 may be responsible for the development of the three avian UV pigments.
Figure 4.
Variation at the eight avian UV pigment-specific amino acid sites. The avian UV pigment-specific amino acids and identical amino acids are boxed.
Importantly, because virtually all SWS1 pigments have S84, the mutation S84C must have occurred in the ancestral avian violet pigment. Thus, if this site is involved in the development of the avian UV pigments, then the UV pigment must have evolved from the violet pigment by a single amino acid replacement. As already noted, according to the “amino acid tree” (Fig. 2), this transition might have occurred only once in the avian lineage.
Molecular Evolution of the Avian UV Pigments: Hypothesis Testing.
We first asked whether we could transform the UV pigment of the zebra finch into a violet pigment by a single mutation, C84S. When this amino acid change was introduced into the zebra finch UV pigment, the mutant pigment achieved a significantly red-shifted λmax value at 397 ± 1 nm (Fig. 1B). Both photobleaching (λmax ≈ 380 nm) and acid denaturation (λmax ≈ 440 nm) spectra show that the observed dark spectrum is generated by the visual pigment (Fig. 1B). Thus, C84S is sufficient to transform the zebra finch UV pigment with a λmax at 359 nm into a violet pigment. Note that the absorption spectrum of the mutant pigment is somewhat broader than that of the wild-type pigment. By subjecting the pigments to various pH conditions, we have attempted to narrow the width of the absorption spectrum of the mutant pigment. However, the mutant pigment at pH 4.4, 4.8, 5.5, 6.2, 6.4, 6.6, 7.2, 7.5, 7.8, 8.6, and 11.4 shows identical absorption spectrum to that at pH 6.6 (results not shown). Thus, the various levels of acid treatment suggest that there is no change in its protonation state that influences the electrostatic environment of the chromophore (26). At present, it is not clear why the absorption spectrum of the mutant pigment is broader than that of the UV pigment. Importantly, however, the λmax value of the zebra finch mutant pigment is not affected by its slightly broader absorption spectrum.
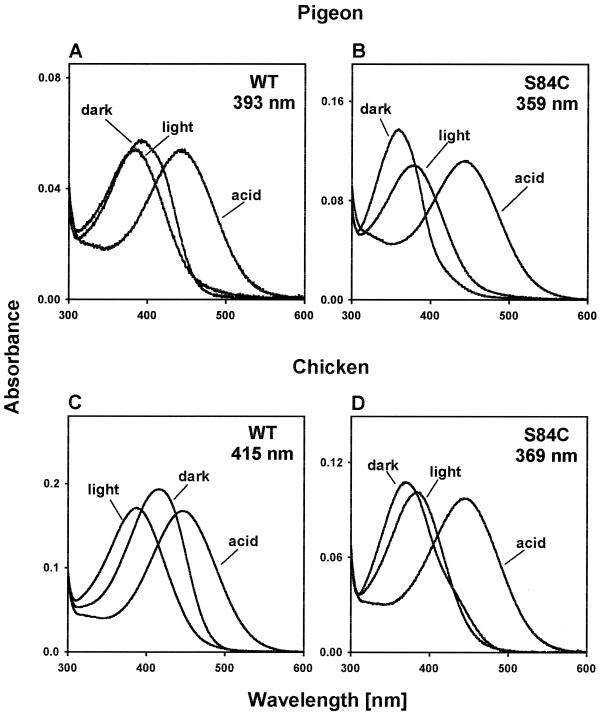
We next attempted to construct UV pigments from the violet pigments of pigeon and chicken by introducing the reverse mutation, S84C. The λmax value of the violet pigment of the pigeon is known to be 393 nm (Fig. 5A; ref. 20). The pigeon pigment with S84C achieves a λmax value at 358 ± 2 nm (Fig. 5B). The photobleaching and acid denaturation experiments again demonstrate that these absorption spectra are generated by visual pigment (Fig. 5 A and B). For the chicken, we had to produce its violet pigment before the mutagenesis experiment (see Materials and Methods). It should be noted that the λmax value at 415 nm (Fig. 5C) of the chicken violet pigment regenerated is virtually identical to the previously estimated λmax values at 413–415 nm using different methods (27–29). The violet chicken pigment with S84C achieves a λmax value at 369 ± 1 nm (Fig. 5D). Again, the photobleaching and acid denaturation experiments show that the dark spectrum is attributable to visual pigments (Fig. 5 C and D). All of these mutagenesis experiments strongly support the hypothesis that the avian UV pigments have evolved from violet pigments by a single amino acid replacement, S84C. Careful inspection of the dark spectra reveals that those of the pigeon violet pigment (Fig. 5A) and the chicken mutant pigment (Fig. 5D) also have slightly broader widths compared with those of the corresponding spectra. Again, the λmax values of these pigments are not affected by the broader distributions.
Figure 5.
Absorption spectra of the pigeon and chicken pigments. (A and B) Absorption spectra of the wild-type (WT) and mutant (S84C) pigments of the pigeon and those after the exposure to light (light) and H2SO4 (acid). (C and D) Absorption spectra of the wild-type and mutant pigments of the chicken and those after the exposure to light and H2SO4. The ratios of the protein absorption peak (not shown) to the pigment absorption peak were 3.59 (A), 2.69 (B), 2.67 (C), and 3.59 (D).
The present analyses show that the dramatic blue-shift in the λmax value of the avian UV pigments is caused simply by the replacement of the hydroxyl group of S84 by the sulfhydryl group of C84. The amino acid site 84 is located near the Schiff base nitrogen and E107 (corresponds to E113 of the bovine rhodopsin), counterion of the Schiff base (12, 26, 30). Using bovine rhodopsin, it has been suggested that one or a few water molecules is located in this region (31–34). Because of its hydrophobicity, it is highly likely that C84 has depleted a water molecule from the avian UV pigments and displaced a positive charge away from the Schiff base (34). Thus, our results strongly suggest that the chromophores of the avian UV pigments are unprotonated, causing a major blue-shift in the λmax value (35, 36). The sulfhydryl group of C84 may also cause further blue-shift in the λmax by forming a disulfide link and modifying the pigment structure.
UV Vision in Birds and Other Vertebrates.
Color vision of birds is affected strongly by colored oil droplets (10, 11). However, the UV and violet photoreceptors of birds contain transparent oil droplets that have no significant light absorption throughout the spectrum, and, consequently, the UV vision of birds is determined solely by the visual pigments (10–12). It is important to note that some bird species can also achieve UV vision without having “true” UV pigments. For example, despite having violet pigments, the pigeon is known for its UV-sensitivity (e.g., see ref. 37). This is possible because the pigeon's cornea, lens, and vitreous body transmit both “visible” and UV light (38). Once the UV light reaches the retina, the pigeon can detect it with the violet pigments that are sensitive to wavelengths between 320 and 450 nm (Fig. 5A). However, UV vision in a much wider variety of avian species is based entirely on the “evolutionarily more specialized” UV pigments (10, 11, 39). Thus, our analyses demonstrate that the origin of the specialized UV vision of many bird species can be traced to a single amino acid change.
Goldfish (P359), zebrafish (P362), chameleon (P358), mouse (P359), and rat (P358) pigments do not have C84 at the corresponding sites (Fig. 4). Evidently, the role of S84C in achieving UV-sensitivity is limited to the avian species, and the molecular bases of UV vision of other vertebrate species are entirely different from that of UV vision of birds. The present evolutionary approach in elucidating the genetic basis of the avian UV vision may also be applied to solve the molecular bases of UV vision in other vertebrates.
Acknowledgments
We thank J. Belote, P. Dunham, R. Yokoyama, and an anonymous reviewer for their comments on the earlier draft. This work was supported by National Institutes of Health Grant GM42379.
Abbreviations
- SWS1
short wavelength-sensitive type 1
- LWS
long wavelength-sensitive
- MWS
middle wavelength-sensitive
- RH1
rhodopsin
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF222331).
References
- 1.Jacobs G H. Am Zool. 1992;32:544–554. [Google Scholar]
- 2.Burkhardt D. Naturwissenschaften. 1982;69:153–157. doi: 10.1007/BF00364887. [DOI] [PubMed] [Google Scholar]
- 3.Burkhardt D. J Comp Physiol A. 1989;164:787–796. [Google Scholar]
- 4.Harosi F I. In: The Visual System. Fein A, Levine J S, editors. New York: Liss; 1985. pp. 41–55. [Google Scholar]
- 5.Fleishman L J, Loew E R, Leal M. Nature (London) 1993;365:397. [Google Scholar]
- 6.Viitala J, Korpimaki E, Palokangas P, Koivula M. Nature (London) 1995;373:425–427. [Google Scholar]
- 7.Bennett A T D, Cuthill I C, Partridge J C. Nature (London) 1996;380:433–435. [Google Scholar]
- 8.Bennett A T D, Cuthill I C, Partridge J C, Lunau K. Proc Natl Acad Sci USA. 1997;94:8618–8621. doi: 10.1073/pnas.94.16.8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunt S, Cuthill I C, Swadle J P, Bennett A T D. Anim Behav. 1997;54:1383–1392. doi: 10.1006/anbe.1997.0540. [DOI] [PubMed] [Google Scholar]
- 10.Goldsmith T H, Collins J S, Licht S. Vision Res. 1984;24:1661–1671. doi: 10.1016/0042-6989(84)90324-9. [DOI] [PubMed] [Google Scholar]
- 11.Bowmaker J K, Heath L A, Wilkie S E, Hunt D M. Vision Res. 1997;16:2183–2194. doi: 10.1016/s0042-6989(97)00026-6. [DOI] [PubMed] [Google Scholar]
- 12.Das D, Wilkie S E, Hunt D M, Bowmaker J K. Vision Res. 1999;39:2801–2815. doi: 10.1016/s0042-6989(99)00023-1. [DOI] [PubMed] [Google Scholar]
- 13.Yokoyama S. Mol Biol Evol. 1994;11:32–39. doi: 10.1093/oxfordjournals.molbev.a040090. [DOI] [PubMed] [Google Scholar]
- 14.Yokoyama S. Mol Biol Evol. 1995;12:53–61. doi: 10.1093/oxfordjournals.molbev.a040190. [DOI] [PubMed] [Google Scholar]
- 15.Yokoyama S. Annu Rev Genet. 1997;31:311–332. doi: 10.1146/annurev.genet.31.1.315. [DOI] [PubMed] [Google Scholar]
- 16.Yokoyama S, Yokoyama R. Annu Rev Ecol Syst. 1996;27:543–567. [Google Scholar]
- 17.Hisatomi O, Satoh T, Barthel L K, Stenkamp D L, Raymond P A, Tokunaga F. Vision Res. 1996;36:933–939. doi: 10.1016/0042-6989(95)00189-1. [DOI] [PubMed] [Google Scholar]
- 18.Wilkie S E, Vissers P M A M, de Grip W J, Bowmaker J K, Hunt D M. Biochem J. 1998;330:541–548. doi: 10.1042/bj3300541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawamura S, Yokoyama S. Vision Res. 1998;38:37–44. doi: 10.1016/s0042-6989(97)00160-0. [DOI] [PubMed] [Google Scholar]
- 20.Yokoyama S, Radlwimmer F B, Kawamura S. FEBS Lett. 1998;423:155–158. doi: 10.1016/s0014-5793(98)00086-6. [DOI] [PubMed] [Google Scholar]
- 21.Yokoyama S. Methods Enzymol. 2000;315:312–325. doi: 10.1016/s0076-6879(00)15851-3. [DOI] [PubMed] [Google Scholar]
- 22.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 23.Felsenstein J. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 24.Kimura M. Proc Natl Acad Sci USA. 1981;78:454–458. doi: 10.1073/pnas.78.1.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar S, Hedges S B. Nature (London) 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 26.Lin S W, Kochenderfer G G, Carroll K S, Wang D, Mathies R A, Sakmar T P. J Biol Chem. 1998;273:24583–24591. doi: 10.1074/jbc.273.38.24583. [DOI] [PubMed] [Google Scholar]
- 27.Govardovskii V I, Zueva l. Vision Res. 1977;17:537–543. doi: 10.1016/0042-6989(77)90052-9. [DOI] [PubMed] [Google Scholar]
- 28.Norren D V. Vision Res. 1975;15:1164–1166. doi: 10.1016/0042-6989(75)90017-6. [DOI] [PubMed] [Google Scholar]
- 29.Okano T, Fukada Y, Artamonov I D, Yoshizawa T. Biochemistry. 1989;28:8848–8856. doi: 10.1021/bi00448a025. [DOI] [PubMed] [Google Scholar]
- 30.Rao V R, Cohen G B, Oprian D D. Nature (London) 1994;367:639–642. doi: 10.1038/367639a0. [DOI] [PubMed] [Google Scholar]
- 31.Deng H, Huang L, Callender R, Ebrey T. Biophys J. 1994;66:1129–1136. doi: 10.1016/S0006-3495(94)80893-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harosi F, Sandorfy C. Photochem Photobiol. 1995;61:510–517. [Google Scholar]
- 33.Nagata T, Terakita A, Kandori H, Kojima D, Shichida Y, Maeda A. Biochemistry. 1997;36:6164–6170. doi: 10.1021/bi962920t. [DOI] [PubMed] [Google Scholar]
- 34.Rafferty C N, Shichi H. Vision Res. 1981;33:229–234. doi: 10.1111/j.1751-1097.1981.tb05329.x. [DOI] [PubMed] [Google Scholar]
- 35.Fasick J I, Lee N, Oprian D D. Biochemistry. 1999;38:11593–11596. doi: 10.1021/bi991600h. [DOI] [PubMed] [Google Scholar]
- 36.Vought B W, Dukkipatti A, Max M, Knox B E, Birge R R. Biochemistry. 1999;38:11287–11297. doi: 10.1021/bi990968b. [DOI] [PubMed] [Google Scholar]
- 37.Kawamura S, Blow N S, Yokoyama S. Genetics. 1999;153:1839–1850. doi: 10.1093/genetics/153.4.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emmerton J, Delius J. Comp Physiol. 1980;141:47–52. [Google Scholar]
- 39.Hart N S, Partridge J C, Cuthill I C. J Exp Biol. 1998;201:1433–1446. doi: 10.1242/jeb.201.9.1433. [DOI] [PubMed] [Google Scholar]
- 40.Hargrave P A, McDowell J H, Curtis D R, Wang J K, E, Juszczak E, Fong S L, Mohanna Rao J K, Argos P. Biophys Struct Mech. 1983;9:235–244. doi: 10.1007/BF00535659. [DOI] [PubMed] [Google Scholar]