Abstract
As the amount of biological data grows, so does the need for biologists to store and access this information in central repositories in a free and unambiguous manner. The European Bioinformatics Institute (EBI) hosts six core databases, which store information on DNA sequences (EMBL-Bank), protein sequences (SWISS-PROT and TrEMBL), protein structure (MSD), whole genomes (Ensembl) and gene expression (ArrayExpress). But just as a cell would be useless if it couldn't transcribe DNA or translate RNA, our resources would be compromised if each existed in isolation. We have therefore developed a range of tools that not only facilitate the deposition and retrieval of biological information, but also allow users to carry out searches that reflect the interconnectedness of biological information. The EBI's databases and tools are all available on our website at www.ebi.ac.uk.
INTRODUCTION: WHAT IS THE EBI?
The roots of the European Bioinformatics Institute (EBI) lie in the European Molecular Laboratory (EMBL) Nucleotide Sequence Data Library (1), which was established in 1980 at the EMBL in Heidelberg, Germany and was the world's first nucleotide sequence database. What began as a modest task of abstracting information from the literature soon became a major database activity with direct electronic submissions of data and the need for highly skilled informatics staff. The task grew with the start of the genome projects and, in 1992, the decision was taken to create a new EMBL Outstation dedicated to bioinformatics. The EBI was established on the Wellcome Trust Genome Campus in the United Kingdom, in close proximity to major sequencing efforts taking place at the Wellcome Trust Sanger Institute and the Human Genome Mapping Project (HGMP) Resource Centre.
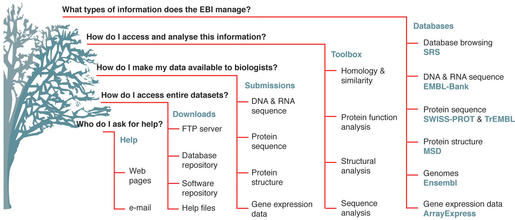
The EBI fulfils three important functions: it provides some of the world's largest and most heavily used bioinformatics services in a free and unrestricted manner; it carries out research; and it trains the bioinformaticians of the future. We will limit our discussion here to the EBI's databases and their services (Fig. 1).
Figure 1.
Summary of the services available at the EBI's website.
DATABASES
The six core molecular databases hosted by the EBI reflect the methods used by biologists to collect information on how cells and organisms work. These store information on DNA and RNA sequences (EMBL-Bank), protein sequences (SWISS-PROT and TrEMBL), protein structure (MSD), whole genomes (Ensembl) and gene expression experiments (ArrayExpress). All the EBI's databases are annotated: features pertaining to gene structure and transcription mechanisms as well as those describing protein structure and function are stored in them and are predicted, inferred, interpreted and validated from many sources. Much of this annotation is performed by highly qualified biologists and the automated annotation that we do is subjected to rigorous quality control. Furthermore, our databases are extensively cross-referenced: our curators have added over 5 million database cross-references. This makes it straightforward for users to determine the relationships that exist between the different types of molecules represented in the EBI's databases, as well as providing easy ways for linking out to resources that are provided by our collaborators at other institutions.
DNA AND RNA SEQUENCES
It would now be impossible for molecular biologists to do their research without free access to nucleotide sequence data. EMBL-Bank is Europe's primary nucleotide sequence resource. It is produced in an international collaboration with GenBank in the USA (2) and the DNA Database of Japan (DDBJ) (3). Each of the three groups collects a proportion of the total sequence data reported worldwide, and all new and updated database entries are exchanged between them on a daily basis.
EMBL-Bank contains tens of millions of DNA and RNA sequences, ranging from as few as ten base pairs to entire genomes. Its sequences come from three main sources: individual research groups, genome-sequencing projects and patent applications. EMBL-Bank is a primary database: the submitters own the data. Although large-scale sequencing projects have now become EMBL-Bank's main source of new sequence data, the importance of short sequences submitted by individual researchers should not be underestimated. These submissions often embody the results of detailed research into gene function and contribute to the body of knowledge to an extent that belies the modest volume of data. EMBL-Bank's curators make sure that this information is up to the high standards of annotation required for their public distribution, and these highly annotated sequences are often updated as more knowledge about them is determined by the original authors (or other contributors; see below). By contrast, genome-sequencing projects are usually long-term collaborations between a sequencing group and EMBL-Bank, resulting in huge amounts of automatically annotated data being submitted directly to the database. Finally, EMBL-Bank has a direct pipeline from the European Patent Office, so all sequences in patent applications are incorporated into EMBL-Bank as soon as they become publicly available.
EMBL-Bank has been growing exponentially for more than ten years now. More than 38% of the total nucleotide count in the database is from the human genome alone. This is followed closely by mouse and rat, which account for another 40%. Owing to their economic importance, plant genomes, particularly that of rice, now form a significant proportion of EMBL-Bank's content, with three different strains available in the database. Model organisms such as the fruit fly and the nematode represent 2% and 3% of total nucleotide count, respectively. Rapid and accurate DNA sequencing has also significantly enhanced the fight against disease. Most of the completed genomes available in EMBL-Bank are from sequencing projects that aim to deal with cancer, hunger (through the plant-genome-sequencing projects) and infectious disease. More than 100 bacterial genomes have been completed in the past two years and EMBL-Bank contains more than 2400 viral genomes.
Accessing EMBL-Bank
The main ways to access EMBL-Bank are through a query tool called the Sequence Retrieval System (SRS; see later) (4), which is available from the EBI at srs.ebi.ac.uk/, or by downloading data from the EBI's FTP server. Data can be retrieved from any of the three collaborating databases, regardless of which one it was submitted to. The EBI also provides a range of tools (Table 1) so that users can run sequence similarity searches against EMBL-Bank. At each major release of the database a DVD set is produced with the compressed content of the database. This set is available on request from support@ebi.ac.uk.
Table 1. A selection of data-access tools available at the EBI.
| Tool | Description |
|---|---|
| Homology and similarity | |
| Fasta and blast tools | Sequence similarity and homology searching against nucleotide and protein databases. Fasta can be very specific when identifying long regions of low similarity, especially for highly diverged sequences. Blast is fast and sensitive, yielding functional and evolutionary clues about the structure and function of your query sequence. |
| Genome and Proteome Fasta3 server | Sequence similarity and homology searching against complete proteome or genome databases using Fasta. |
| SNP-Fasta3 server | Fasta3 searches of the European database of single nucleotide polymorphisms (HGVbase). |
| MPsrch | Aneda's very fast implementation of the true Smith and Waterman algorithm for biological sequence comparison. MPsrch identifies hits often missed by other database-searching methods. |
| Scanps2.3 | Scan Protein Sequence: fast implementation of the true Smith and Waterman algorithm for protein database searches. |
| Sequence analysis | |
| ClustalW | Produces biologically meaningful multiple sequence alignments of divergent sequences. |
| GeneMark | A gene-prediction service that assesses the coding potential of DNA sequences by using Markov models of coding and non-coding regions within a sliding window. |
| Genetic Code Viewer | Highlights genetic code differences in different organisms. |
| Protein functional analysis | |
| CluSTr Search | Search the EBI's protein sequence databases (SWISS-PROT and TrEMBL) by accession numbers. |
| InterProScan | Compare your query protein sequence with those in InterPro—the EBI's resource for protein families, domains and functional sites. |
| GeneQuiz | Highly automated analysis of biological sequences. |
| Protein structural analysis | |
| DALI | A network service for comparing three-dimensional protein structures. |
| PQS | Search the list of likely quaternary structures generated at the EBI. |
| ChemPDB | A ligand search service: query the MSD's ligands and small molecule dictionary for small molecules or substructures. |
| SSM | Secondary structure matching: compare protein chains/structures and look for similar ones in the whole PDB archive or among SCOP domains. |
| Gene expression analysis | |
| Expression Profiler | A set of tools for clustering, analysing and visualizing gene expression and other genomic data. |
The EBI's Genome Webserver—www.ebi.ac.uk/genomes/—allows users to browse all the completed genomes that have been submitted to EMBL-Bank. Unfinished genomic data can also be accessed using the EBI's Genome Monitoring Table (Genome MOT) (5), in which the data are sorted by chromosome. Genome MOT is updated daily and provides direct access to individual EMBL-Bank entries.
Submitting data to EMBL-Bank
Most journals now require authors to submit sequence information to EMBL-Bank, GenBank or DDBJ before publication. This maximizes its benefit to research, by ensuring that it will always be available and readily accessible to scientists. EMBL-Bank assigns each submitted sequence an accession number that permanently identifies it, and that authors include when they publish a sequence. This procedure ensures availability and distribution of new sequence data in a timely fashion. Upon submission, authors can either make their sequence publicly available immediately or wait until their paper is published.
Webin (Table 2), EMBL-Bank's submission system, guides the user through a sequence of WWW forms allowing interactive submission of sequence data and descriptive information (such as coding and regulatory regions). All the information required to create a database entry is collected during this process. Webin is designed to allow fast submission of single, multiple or very large numbers of sequences (bulk submissions) and is available at www.ebi.ac.uk/embl/Submission.
Table 2. Submission tools for the EBI's core databases.
| Submission tool | Description |
|---|---|
| Nucleotide sequence submissions | |
| Webin | Interactive system for submitting DNA sequences to the EMBL-Bank, GenBank or DDBJ sequence databases. You can also report updates or corrections for existing EMBL-Bank nucleotide sequence entries and submit multiple sequence alignments. |
| Genome Project Accounts | Large volumes of genome sequence data can be deposited and updated by the originating group via FTP or email. |
| Protein sequence submissions | |
| SWISS-PROT | Submit protein sequences to SWISS-PROT and submit corrections to an existing (publicly available) SWISS-PROT entry. |
| Protein structure submissions | |
| PDB-AutoDep | Three-dimensional structure submissions to the Protein Data Bank. |
| EM-Dep | Submit three-dimensional electron microscopy data to the MSD electron microscopy database. |
| Gene expression data submissions | |
| MIAMExpress | MIAME-compliant microarray data submissions to the ArrayExpress database. |
The EBI works closely with genome-sequencing centres to ensure the timely incorporation of their data into EMBL-Bank for public release. EBI biologists regularly communicate with the sequencing groups to optimize the acquisition and release of sequencing data and the descriptive information associated with them.
Updating existing database entries
Over time, an entry that was correct when it was created can become out of date: authors might make corrections to the sequence itself, or might discover new features to be added to the annotation. Because such findings are rarely published in journals, it is important that authors communicate them to EMBL-Bank. A new feature implemented in 2002 is that researchers can now improve entries submitted by others: the database collaboration with GenBank and the DDBJ has created a dataset into which third-party annotation (TPA) can be deposited, so long as it is supported by publication in a peer-reviewed journal. Updates can be sent to EMBL-Bank using the form at www3.ebi.ac.uk/Services/webin/update/update.html.
PROTEIN SEQUENCES AND FUNCTIONAL INFORMATION
The path from genes (in EMBL-Bank) to their products is far from simple. Alternative splicing, RNA editing and post-translational modifications all increase the ratio of gene products to genes. As the sequences of many genomes near completion, researchers are focusing their efforts on collecting information on all the proteins encoded by them. Protein modifications are well characterized on a protein-by-protein basis but very little is known in a genome-wide context. The EBI strongly believes that rigorously maintained efforts to predict and annotate proteins are central to our understanding of protein variation and function.
The SWISS-PROT Protein Knowledgebase (6,7)—which is maintained collaboratively by the EBI and the Swiss Institute of Bioinformatics (SIB)—provides thorough descriptions of a non-redundant set of proteins, including function, domain structure, post-translational modifications, and variants (5). SWISS-PROT is mainly a secondary database in that most of its content is derived from primary sources such as EMBL-Bank. It is thus tightly integrated with other databases and is extensively cross-referenced. SWISS-PROT's unparalleled level of annotation is possible because it is manually curated. However, this high level of annotation has also slowed the growth of SWISS-PROT. The annotation bottleneck is partially addressed by TrEMBL (Translated EMBL) (7)—the computer-annotated protein sequence database that the EBI produces to supplement SWISS-PROT. TrEMBL contains the translations of all the coding sequences in EMBL-Bank, DDBJ and GenBank, except those already in SWISS-PROT. TrEMBL also contains experimentally determined protein sequences from the literature and those that have been submitted directly to SWISS-PROT. Sequences in TrEMBL are automatically annotated according to a set of more than 500 rules, which exploit protein motifs and annotation from similar sequences. This automatic annotation is always implemented with a human ‘sanity check’. This pipeline brings the annotation standard of TrEMBL closer to that of SWISS-PROT.
SWISS-PROT has grown in a near linear fashion since its first release in 1986. TrEMBL, on the other hand, has grown exponentially since 1996 when the project started. The genome-sequencing projects are the main contributors to this growth. Complete prokaryotic proteomes represent 25% of TrEMBL content, whereas eukaryotic gene products, including human, mouse, fruit fly, nematode and cress, represent 10% at present.
A complete non-redundant protein sequence collection derived from SWISS-PROT and TrEMBL is available, called SPTR (Swall). This allows the user to obtain all the relevant information on a protein or group of proteins in one search. In addition, SPTR offers access to the annotation improvements and updates that the SWISS-PROT and TrEMBL teams make between releases, as SPTR is produced on a weekly basis.
Accessing SWISS-PROT and TrEMBL
The most effective way to browse through SPTR is to use SRS (see later) (4). Documentation can be found at www.ebi.ac.uk/swissprot and www.ebi.ac.uk/trembl. SWISS-PROT, TrEMBL, TrEMBLnew (all the new entries in TrEMBL produced since the last release) and SPTR are also available by anonymous FTP from ftp://ftp.ebi.ac.uk/databases (see below). Finally, SWISS-PROT and TrEMBL full releases are distributed on CD-ROM by the EBI. These are available upon request from support@ebi.ac.uk.
Submitting data to SWISS-PROT and TrEMBL
SWISS-PROT provides accession numbers only for proteins that have been directly sequenced. We do not provide, in advance, accession numbers for protein sequences that are the result of translation of nucleic acid sequences. These translations are automatically forwarded to us from the EMBL-Bank, GenBank and DDBJ nucleotide sequence databases, and are processed into TrEMBL at each release. The SWISS-PROT data submission form (Table 2) is available at www.ebi.ac.uk/swissprot/Submissions.
A ONE-STOP SHOP FOR PROTEIN DOMAIN INFORMATION
InterPro (8) is an integrated documentation resource for protein families, domains and functional sites. InterPro amalgamates a number of databases that use different methodologies and varying degrees of biological information to derive protein signatures from well-characterized proteins. The member databases include Prosite (9), Prints (10), ProDom (11), Pfam (12), SMART (13) and TIGRFAMs (14), which contribute patterns and profiles, fingerprints, clustered domains, and hidden Markov models, respectively. New data from the member databases are integrated from the protein signature databases just before their releases, and all matches against SWISS-PROT and TrEMBL are calculated weekly. The annotation for InterPro entries is derived from merged annotation taken from the member databases, with additional help from experts in the field. Where protein signatures from different member databases describe the same biological protein family or domain, they are united into a single InterPro entry, which contains annotation and a list of all the proteins in SWISS-PROT and TrEMBL that match the signatures in the entry. By uniting the member databases, InterPro capitalizes on their individual strengths, producing a powerful integrated tool for protein sequence classification.
InterPro currently contains information on more than 10 000 unique protein function criteria, which are condensed into more than 5800 distinct InterPro entries. Eighty-five percent of SWISS-PROT and 75% of TrEMBL entries have one or more matches in InterPro.
Retrieving data from InterPro
Interpro is available for text- and sequence-based searching via a webserver at www.ebi.ac.uk/interpro. There is a simple text search as well as an SRS-based text search available for more complex queries involving InterPro, SWISS-PROT and TrEMBL. InterProScan (15) is a sequence-search tool that combines the individual search methods of the member databases into a single package. InterProScan is also available via a mailserver. The InterPro data and a standalone, Perl-based version of InterProScan are also available via anonymous FTP at ftp://ftp.ebi.ac.uk/pub/databases/interpro. This allows local installation of InterProScan, to speed up bulk searches and allow confidential searching of the InterPro database. Additional files available on the FTP site include a list of all InterPro entries, a file of the InterPro to Gene Ontology (GO) mappings (see below) and a list of all hits in SWISS-PROT and TrEMBL. The database and matches are available in XML format with a corresponding document type definition (DTD) file.
PROTEINS IN THREE DIMENSIONS
The Macromolecular Structure Database (MSD) (16) is the European project for the collection, management and distribution of data about macromolecular structures. MSD is a relational database that presents the Protein Data Bank's three-dimensional data in a consistent way. The main database of structure coordinates at the EBI, MSD/PDB, has almost 19 000 entries, including viruses, proteins and peptides, protein–nucleic acid complexes and DNA/RNA structures. Fewer than 4000 entries in SPTR contain links to the PDB. However, more than 90% contain multiple references to distinct PDB structures. Eighty percent of all structures have been determined by X-ray diffraction and 15% from NMR. There are 2.6% theoretical models.
MSD also produces and provides access to databases that support and enrich the structure coordinates set. One example is the Ligand Library (ChemPDB) (16), which contains the comprehensive structural descriptions of all chemical components in the PDB and allows users to search for ligands on the basis of chemical substructures. The MSD group also provides access to secondary-structure-matching services such as SSM (16) and DALI (17), as well as searches on quaternary structure predictions that are generated at the EBI.
Accessing MSD
The simplest way to search MSD/PDB is using the OCA browser available at oca.ebi.ac.uk, which integrates PDB with a range of resources including Pfam (Protein Families), SCOP (Structural Classification of Proteins), KEGG (Kyoto Encyclopedia of Genes and Genomes), GPCRDB (a database of G-protein-coupled receptors) and PubMed references. This allows the user to search using criteria such as PDB ID, keyword, author or method. PDB and several other related databases are also available through the EBI's SRS server (4) in special library sections for secondary and 3D structures.
Submitting data to MSD
MSD runs a deposition service (Table 2) that allows authors to submit crystallographic or nuclear magnetic resonance spectroscopy data to the PDB through AutoDep (www.ebi.ac.uk/msd-srv/autodep) (18). A new service, EM-Dep (www.ebi.ac.uk/msd-srv/emdep), allows users to submit three-dimensional electron microscopy data to EMD, MSD's electron microscopy database (Table 2).
EXPLORING WHOLE GENOMES
Each of the databases described above is largely based around individual genes or their products, but genomics now allows us to take a more holistic approach to biology. Ensembl (19) was initially designed to make the assembled human genome, automatically annotated to a consistent standard, readily accessible to biologists. It is rapidly becoming a Noah's Ark for metazoan genomes, and now includes those of mouse, zebrafish, pufferfish and mosquito. The main aims of Ensembl are to provide scientific content to draft genomic sequence in a fast and reliable manner. Ensembl is produced in a collaboration between the EBI and the Wellcome Trust Sanger Institute.
Accessing Ensembl
Ensembl is available as an interactive website at www.ensembl.org/. Several views of the data are available, allowing the user to zoom in from entire chromosomes (contigview) to genes (geneview) and the proteins encoded by them (proteinview). The user can customize which features to browse in each of these views. Similarity searches [BLAST (20) and SSAHA (21)] are integrated into the Ensembl web browser. Ensembl data sets can also be searched using SRS. Ensembl uses the distributed annotation system (DAS, www.biodas.org) (22), which allows third parties to create local annotation that can be made visible by other users on demand.
Ensembl's data sets and the entire Ensembl software system can be downloaded from www.ensembl.org/Docs/, allowing users to process their own data using Ensembl. The data sets are available in several standard database formats.
TRANSCRIPTOME ANALYSIS
Microarrays are one of the most important recent breakthroughs in the experimental life sciences (23). They allow snapshots to be made of gene expression levels on a genomic scale, and are revolutionizing all areas of the molecular life sciences, from basic biology to drug discovery. The international Microarray Gene Expression Data (MGED) Society (www.mged.org) is a grass roots movement whose aim is to develop standards for microarray experiment annotation—the Minimum Information About a Microarray Experiment (MIAME) (24) and data representation and exchange—Microarray Gene Expression Markup Language (MAGE-ML) (25). Describing microarray experiments and the data generated from them in standardized ways facilitates the sharing of these data. Furthermore, it provides a solid infrastructure for the dissemination of experimental results and the protocols that led to them. ArrayExpress (26) is a new public repository that aims to store well-annotated gene expression data in accordance with MIAME recommendations.
Accessing ArrayExpress
ArrayExpress can be browsed and searched from www.ebi.ac.uk/arrayexpress. Data query, annotation and analysis tools are being developed, and work is under way to populate ArrayExpress with microarray data, array designs and protocols. Over the next year, we expect that the amount of data available in ArrayExpress will grow considerably. The driving force for this will be the requirement for journals to release data on publication, and the Nature journals now require submission of data to a public repository such as ArrayExpress on publication (27).
Submitting data to ArrayExpress
ArrayExpress accepts three different types of submissions: experiments, array designs and protocols. MGED developed MAGE-ML (25) for the description of microarray data. ArrayExpress accepts data either directly in MAGE-ML or via a data-submission tool called MIAMExpress (Table 2), which allows users to submit all three types of data in a MIAME-compliant format.
CONTROLLED VOCABULARIES TO DESCRIBE BIOLOGY
The GO project (28,29) is a collaborative effort to address the need for consistent descriptions of gene products in different databases. The GO collaborators, based in several locations worldwide, are developing three structured, controlled vocabularies (ontologies) that describe biological processes (e.g. protein biosynthesis; ribosome assembly), cellular components (e.g. nucleolus; ribosome) and molecular functions (e.g. aminoacyl tRNA synthetase; translation elongation factor) in a species-independent manner. The EBI is responsible for two aspects of the GO project: creating and editing the ontology terms (in collaboration with other members of the GO consortium), and annotating several of the EBI's databases with GO terms (35).
GO terms do not describe gene products themselves, but the way that they behave in a cellular context. For example, cytochrome is not in the molecular function ontology, but the functions that cytochromes perform, such as electron transporter, are. Gene products are assigned to GO terms through the GO annotation project (GOA), as well as through the efforts of the model organism databases that are members of the GO consortium. GO terms are being applied to a non-redundant set of proteins described in the EBI's core genome and proteome databases (SWISS-PROT, TrEMBL and Ensembl) that collectively provide complete proteomes for humans and other organisms.
The GOA project has assigned GO terms to all the complete and incomplete proteomes that exist in SWISS-PROT and TrEMBL, using a combination of electronic mappings and manual curation. GOA is updated on a monthly basis, in accordance with the latest data released by SWISS-PROT, TrEMBL, Ensembl and InterPro. By annotating all characterized proteins with GO terms and facilitating the transfer of this knowledge to similar uncharacterized proteins, the GOA project has made a significant contribution to providing protein functional information to biologists and bioinformaticians, and is facilitating the integration of data among the EBIs databases.
Accessing GO and GOA
Several browsers are available that allow users to explore the ontologies and find out which genes have been annotated to them. These are available at www.geneontology.org/#tools, and it would be beyond the scope of this review to describe them here. The GO files can also be downloaded from www.geneontology.org/#ftp in three different formats: flat files (updated daily), XML (updated monthly) and MySQL (updated monthly).
GOA project data can be accessed and searched using QuickGO, a fast web-based browser that provides access to core GO data and up-to-date electronic and manual EBI GO annotations. The GOA database can be searched using the EBI's SRS server (4).
The GOA files (in flat-file format) can also be downloaded from www.ebi.ac.uk/GOA/. Two GOA association files (tab-delimited files of associations between gene products and GO terms) are currently produced: the human GOA file contains GO annotations for all proteins in the nonredundant human proteome set; the SPTR GOA file contains GO annotations for all proteins in SPTR. A file of cross references that displays the relationship between the entries in the GOA data set with other databases, such as the nucleotide sequence databases, HUGO, LocusLink and Refseq, is also available.
SEARCHING ACROSS MULTIPLE DATABASES
The SRS (4), initially created and distributed by EMBL/EBI and now produced by Lion Bioscience (www.lionbioscience.com/), is freely available for use at the EBI (srs.ebi.ac.uk/). SRS integrates and links a comprehensive collection of databases, including many of the EBI's core databases. SRS allows users to navigate between databases and supports complex queries. For example, users can ask SRS to find all the kinases in the human genome that have been implicated in cancer and for which the three-dimensional structure has been solved. SRS exploits the fact that many of its component databases contain explicit cross-reference information, so it can provide accurate links between them. It can also generate implicit links between them to optimally provide content for complex queries.
The EBI's SRS server currently has more than 150 data-bases comprising more than 42 million unique records of information. The system also provides access to mainstream bioinformatics applications for sequence database searching and protein and nucleic acid sequence analysis. This includes more than 100 applications from the EMBOSS (30) project, of which the EBI is a collaborator.
BULK ACCESS TO EBI DATA
Users who wish to download entire data sets, so that they can build them into their own bioinformatics resources or search them locally, can do so via the EBI's FTP server at ftp://ftp.ebi.ac.uk/. Many of the EBI's datasets are available in several formats, including flat-files, fasta sequence files and XML.
THE EBI TOOLBOX
Biology is not just about accumulating data; to understand how organisms function, researchers need to analyse and manipulate biological information. The EBI's toolbox, available from www.ebi.ac.uk/Tools/, helps users to find relevant information and analyse it efficiently. The toolbox is divided into five compartments. Homology and similarity searches provides mainstream algorithms such as fasta (31), blast (19,32) and the rigorous Smith and Waterman algorithm (30). Through these, users can access all the EBI's sequence databases including EMBL-Bank, SPTR and completed genomes and proteomes. Protein function analysis is mainly encompassed by InterProScan (15), but all the individual methods to search the InterPro member databases are also available. The sequence analysis tools include programmes for sequence alignment, gene prediction, mutation detection and validation. For structural analysis there are tools for secondary and tertiary structure matching such as DALI (17), MaxSprout (34), ChemPDB (16) and SSM (16). Finally, the miscellaneous tools section includes access points to general services as well as the EBI's database-submission tools (Table 2). A selection of the EBI's most popular tools is provided in Table 1.
TOWARDS GLOBAL INTEGRATION
What types of information are biologists likely to need in the future, and what types of resources are we building to allow them to access such information? This issue of Nucleic Acids Research testifies to the proliferation of bioinformatics resources over the past 7 years. Biologists now have the individual data sets to hand, but how do they go about integrating them, so that they can get to grips with how molecules, pathways and entire systems interact to build functional organisms? One crucial factor is the development of global standards to describe biological information. Through extensive collaborations with organizations such as MGED, the GO consortium and the Human Proteomics Organization (HUPO), the EBI is helping to drive the development of global standards for microarray data, functional annotation and proteomics. A second challenge is to develop computational solutions that will allow biologists to query numerous data sources in meaningful ways. Research into using grid-based technologies for this purpose is already under way at the EBI. By continuing to develop these tools and standards, we hope to fulfil our mission to make biological information freely accessible to all facets of the scientific community, in ways that promote scientific progress.
Acknowledgments
ACKNOWLEDGEMENTS
Projects within the EBI are supported by the European Molecular Biology Laboratory (EMBL), the European Commission, the Wellcome Trust, the UK Medical Research Council, the UK Biotechnology and Biosciences Research Council, the UK Engineering and Physical Sciences Research Council and the EBI Industry Programme.
REFERENCES
- 1.Stoesser G. et al. (2002) The EMBL Nucleotide Sequence Database. Nucleic Acids Res., 30, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dennis A. et al. (2002) GenBank. Nucleic Acids Res., 30, 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tateno Y. et al. (2002) DNA Data Bank of Japan (DDBJ) for genome scale research in life science. Nucleic Acids Res., 30, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zdobnov E.M. et al. (2002) The EBI SRS server—new features. Bioinformatics, 18, 1149–1150. [DOI] [PubMed] [Google Scholar]
- 5.Beck S. and Sterk,P. (1998) Genome-scale DNA sequencing: where are we? Curr. Opin. Biotechnol., 9, 116–120. [DOI] [PubMed] [Google Scholar]
- 6.Apweiler R. (2001) Functional annotation in SWISS-PROT: The basis for large-scale characterization of protein sequences. Brief Bioinform., 2, 9–18. [DOI] [PubMed] [Google Scholar]
- 7.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Apweiler R., Attwood,T.K., Bairoch,A., Bateman,A., Birney,E., Biswas,M., Bucher,P., Cerutti,L., Corpet,F., Croning,M.D., Durbin,R., Falquet,L., Fleischmann,W., Gouzy,J., Hermjakob,H., Hulo,N., Jonassen,I., Kahn,D., Kanapin,A., Karavidopoulou,Y., Lopez,R., Marx,B., Mulder,N.J., Oinn,T.M., Pagni,M. and Servant,F. (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res., 29, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falquet L. et al. (2002) The PROSITE database, its status in 2002. Nucleic Acids Res., 30, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Attwood T.K., Blythe,M.J., Flower,D.R., Gaulton,A., Mabey,J.E., Maudling,N., McGregor,L., Mitchell,A.L., Moulton,G., Paine,K. and Scordis,P. (2002) PRINTS and PRINTS-S shed light on protein ancestry. Nucleic Acids Res., 30, 239–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corpet F., Servant,F., Gouzy,J. and Kahn,D. (2000) ProDom and ProDom-CG: tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res., 28, 267–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L.L. (2002) The Pfam Protein Families Database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Letunic I. et al. (2002) Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res., 30, 242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haft D.H., Loftus,B.J., Richardson,D.L., Yang,F., Eisen,J.A., Paulsen,I.T. and White,O. (2001) TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res., 29, 41–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zdobnov E.M. and Apweiler,R. (2001) InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics, 17, 847–848. [DOI] [PubMed] [Google Scholar]
- 16.Boutselakis H. et al. (2003) E-MSD: the European Bioinformatics Institute Macromolecular Structure Database. Nucleic Acids Res., 31, 458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holm L. and Sander,C. (1998) Touring protein fold space with Dali/FSSP. Nucleic Acids Res., 26, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin D. et al. (2000) Autodep: a web-based system for deposition and validation of macromolecular structural information. Acta Crystallogr., D56, 828–841. [DOI] [PubMed] [Google Scholar]
- 19.Hubbard T. et al. (2001) The Ensembl genome database project. Nucleic Acids Res., 30, 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul S.F. et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ning Z. et al. (2001) SSAHA: a fast search method for large DNA databases. Genome Res., 11, 1725–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dowell R.D. et al. (2001) The Distributed Annotation System. BMC Bioinformatics, 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The Chipping Forecast (1999) Nature Genet., 21(Suppl.), 1–60. [Google Scholar]
- 24.Brazma A. et al. (2001) Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nature Genet., 29, 365–371. [DOI] [PubMed] [Google Scholar]
- 25.Spellman P.T. et al. (2002) Design and implementation of microarray gene expression markup language (MAGE-ML). Genome Biol., 3, research0046.1–0046.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brazma A. et al. (2003) ArrayExpress — a public repository for microarray gene expression data. Nucleic Acids Res., 31, 68–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anon. (2002) Microarray standards at last. Nature, 419, 323. [DOI] [PubMed] [Google Scholar]
- 28.Ashburner M. et al. (2000) Gene Ontology: tool for the unification of biology. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashburner M. et al. (2001) Creating the Gene Ontology resource: design and implementation. Genome Res., 11, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rice P., Longden,I. and Bleasby,A. (2000) EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet., 16, 276–277. [DOI] [PubMed] [Google Scholar]
- 31.Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul S.F. and Gish,W. (1996) Local alignment statistics. Methods Enzymol., 266, 460–480. [DOI] [PubMed] [Google Scholar]
- 33.Smith T.F. and Waterman,M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 34.Holm L. and Sander,C. (1991) Database algorithm for generating protein backbone and side-chain co-ordinates from a C alpha trace application to model building and detection of co-ordinate errors. J. Mol. Biol., 218, 183–194. [DOI] [PubMed] [Google Scholar]
- 35.Camon E., Barrell,D., Brocksbank,C., Magrane,M. and Aptveiler,R. The gene ontology annotation project–application of GO in SWISS-PROT, TrEMBL and InterPro. Comparative and Functional Genomics, in press. [DOI] [PMC free article] [PubMed]