Abstract
The Protein Data Bank (PDB; http://www.pdb.org/) continues to be actively involved in various aspects of the informatics of structural genomics projects—developing and maintaining the Target Registration Database (TargetDB), organizing data dictionaries that will define the specification for the exchange and deposition of data with the structural genomics centers and creating software tools to capture data from standard structure determination applications.
INTRODUCTION
Over the history of the Protein Data Bank (PDB; http://www.pdb.org/) (1,2), this archive of three dimensional structural data has grown from 7 files in 1971 to a database containing over 18 800 structures as of October 2002. The archive's growth has been accompanied by increases in both data content and the structural complexity of individual entries. A further acceleration is anticipated due to development in high-throughput structural determination methodologies and worldwide structural genomics efforts.
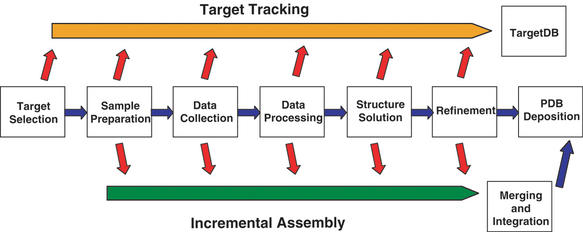
The PDB has been actively involved in various aspects of structural genomics (3), ranging from target tracking to the automation of data deposition from the many steps involved in high throughput structure determination (Fig. 1).
Figure 1.
Schematic diagram of structure determination data pipeline.
TARGET REGISTRATION
Efficient structure solution on a genomic scale requires a centralized coordination of effort, to which the timely availability of status information on the progress of protein production and structure solution is key. Building on the work of earlier target databases [Presage (4) and http://www.genome3d.org/], we have created a centralized target registration database for sequences from worldwide structural genomics projects (TargetDB; http://targetdb.pdb.org/). Target sequences are collected weekly from the P50 NIH structural genomics centers and other international projects (see http://www.rcsb.org/pdb/strucgen.html). Target data are organized following recommendations from the International Task Force on Target Tracking, which include the definitions for the states used to track target progress (5). These states span the details of protein production, structure solution and the ultimate deposition of experimental and structure data. Target data from all contributing structural genomics sites are combined into a single downloadable XML document following the document type definition at http://targetdb.pdb.org/apps/target.txt.
Target sequences are loaded into a relational database, along with the sequences from experimentally determined structures in the PDB (∼41 000 sequences), and with the sequences data depositors have approved for pre-release. From this latter set, sequence information is currently available for about 45% of on-hold depositions.
All or subsets of these sequence data may be searched using the FASTA sequence comparison method (6). A simple search form is provided to permit queries of each target data element, including: contributing site, protein name, sequence, project tracking identifier, date of last modification, current status of the target and source organism. Search results may be viewed as HTML reports, FASTA files or XML documents. TargetDB also tracks target status temporally so reports of the evolution of target progress over a selected time interval can be created.
Table 1 summarizes the status of sequences under study by the structural genomics projects. It is expected that a significant fraction of the thousands of proteins currently in process will ultimately be solved and deposited in the PDB.
Table 1. Breakdown of structural genomics targets by target status.
| Target status | Definition | Number |
|---|---|---|
| Selected | Target selected | 16 822 |
| Cloned | Protein cloned | 9721 |
| Expressed | Protein expressed | 5206 |
| Soluble | Soluble protein | 1988 |
| Purified | Protein purified | 2495 |
| Crystallized | Protein crystals obtained | 1055 |
| Diffraction-quality crystals | Diffraction-quality crystals obtained | 324 |
| Diffraction | Diffraction data collected | 287 |
| HSQC | Heteronuclear Single Quantum Correlation spectra | 235 |
| NMR assigned | NMR spectrum assigned | 72 |
| Crystal structure | Crystal structure obtained | 192 |
| NMR structure | NMR structure obtained | 51 |
| In the PDB | Structure has been deposited to the PDB | 185 |
TargetDB can be queried by the Target Status stages given above. Numbers given show the state of the database as of 12 September 2002.
DATA DICTIONARIES
The International Task Force on Deposition, Archiving and Curation of the Primary Information has recommended that all depositions from structural genomics efforts include information that would normally be found in a Materials and Methods section of a journal article reporting structure determination (7). This has required the addition of new data items in a PDB file which are documented in the PDB exchange data dictionary (http://deposit.pdb.org/mmcif/). This data dictionary expands upon the macromolecular Crystallographic Information File (mmCIF) data dictionary (8), an ontology of data definitions that electronically encodes domain information in the form of precise definitions, examples and controlled vocabularies (9). In addition to domain information, data definitions also encode information such as data type, data relationships, range restrictions, controlled vocabularies and presentation units.
The PDB exchange data dictionary is virtually complete for crystallographic and NMR structure determination and refinement; a protein production dictionary is under development and should be complete in 2003.
The use of software accessible data dictionaries is the key ingredient of the PDB informatics infrastructure (1). The dictionary provides the foundation for software tools used to exchange and validate data, create and load databases, translate data formats and serve application program interfaces.
SOFTWARE TOOLS FOR DATA EXTRACTION AND DEPOSITION
One goal of high-throughput structural genomics is the automatic capture of the important details of each step in the structure determination pipeline. Figure 1 shows the steps in a simplified structure determination data pipeline. At each step, essential details are captured and assembled to make a data file for PDB deposition. The status for each target sequence is updated at each step and forwarded to TargetDB. The PDB data processing system has been developed in anticipation of a structure determination data pipeline with automated deposition as a final step.
The AutoDep Input Tool (ADIT) was originally developed by the PDB to support the centralized data deposition and annotation of macromolecular structure data; however, this system can also support data from the structure determination pipeline. ADIT depends entirely on an underlying data dictionary to define the content and properties of information to be processed. This design permits the system to easily adapt to content extensions without software change. ADIT has been packaged in a workstation mode to provide single user data input and processing functionality tailored specifically for the content requirements of structural genomics applications.
ADIT can capture and edit data files stored in a standard form such as mmCIF and can be used to manually include details that are not captured automatically from a standard format. Although this is a common practice for current PDB depositions, it would obviously be more efficient if all of the information to be captured conformed to a standard data dictionary and format. This standardization is a key requirement for building a robust automated data pipeline.
The PDB exchange data dictionary definitions have been carefully developed to describe the information to be extracted from each step in the structure determination pipeline. The majority of these details are output by structure determination applications, but are not currently produced in a common format. Some applications export information directly in mmCIF format following the exchange dictionary; others produce output in program-specific formats or in a program log file. For the latter, the utility program PDB_EXTRACT was created to extract key data values from common structure determination applications output. PDB_EXTRACT also facilitates the merging of the incremental extractions of data from each program step. In the end, PDB_EXTRACT produces an integrated mmCIF data file that can be imported into ADIT to prepare and check the data file for PDB deposition.
The impact of providing precise data specifications and software tools to depositors is already having an impact on the efficiency of data deposition and annotation. In our first test of fully automated deposition with a NIH P50 structural genomics center (the Joint Center for Structural Genomics), we were able to reduce the total data processing and annotation time for a structure by a factor of 10. As automated deposition technology spreads to other projects inside and outside the structural genomics arena, the PDB will begin to realize the significant benefit of this investment in infrastructure.
ADIT, PDB_EXTRACT and mmCIF parsing and data management tools are currently distributed by the PDB under an open-source license at http://deposit.pdb.org/software/.
FUTURE
The PDB will continue to work with the community to identify and define the required data items for structural genomics and to work with software developers to directly export these data in a common form and integrate this output with the PDB deposition software. Our efforts to produce tools to facilitate the extraction and integration of the data from existing structure determination software will continue. We will further encourage the structure genomics centers to use PDB software tools in their respective data processing operations.
Questions and comments about the PDB should be sent to info@rcsb.org.
Acknowledgments
ACKNOWLEDGEMENTS
The PDB is operated by Rutgers, The State University of New Jersey; The San Diego Supercomputer Center at the University of California, San Diego; and the National Institute of Standards and Technology—three members of the Research Collaboratory for Structural Bioinformatics (RCSB). This work is supported by grants from the National Science Foundation, the Department of Energy, and two units of the National Institutes of Health: the National Institute of General Medical Sciences and the National Library of Medicine.
REFERENCES
- 1.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein F.C., Koetzle,T.F., Williams,G.J., Meyer,E.E., Brice,M.D., Rodgers,J.R., Kennard,O., Shimanouchi,T. and Tasumi,M. (1977) Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol., 112, 535–542. [DOI] [PubMed] [Google Scholar]
- 3.Berman H.M., Bhat,T.N., Bourne,P.E., Feng,Z., Gilliland,G., Weissig,H. and Westbrook,J. (2000) The Protein Data Bank and the challenge of structural genomics. Nature Struct. Biol., 7, 957–959. [DOI] [PubMed] [Google Scholar]
- 4.Brenner S.E., Barken,D. and Levitt,M. (1999) The Presage database for structural genomics. Nucleic Acids Res., 27, 251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Task Force on Target Tracking (2001) Task Force Reports from the Second International Structural Genomics Meeting. Airlie, VA. http://www.nigms.nih.gov/news/reports/airlie_tasks.html.
- 6.Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 24, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Task Force on the Deposition, Annotation, and Curation of the Primary Information (2001) Task Force Reports from the Second International Structural Genomics Meeting. Airlie, VA. http://www.nigms.nih.gov/news/reports/airlie_tasks.html.
- 8.Bourne P.E., Berman,H.M., Watenpaugh,K., Westbrook,J.D. and Fitzgerald,P.M.D. (1997) The macromolecular Crystallographic Information File (mmCIF). Methods Enzymol., 277, 571–590. [DOI] [PubMed] [Google Scholar]
- 9.Westbrook J. and Bourne,P.E. (2000) STAR/mmCIF: An extensive ontology for macromolecular structure and beyond. Bioinformatics, 16, 159–168. [DOI] [PubMed] [Google Scholar]