Abstract
ToxoDB (http://ToxoDB.org) provides a genome resource for the protozoan parasite Toxoplasma gondii. Several sequencing projects devoted to T. gondii have been completed or are in progress: an EST project (http://genome.wustl.edu/est/index.php?toxoplasma=1), a BAC clone end-sequencing project (http://www.sanger.ac.uk/Projects/T_gondii/) and an 8X random shotgun genomic sequencing project (http://www.tigr.org/tdb/e2k1/tga1/). ToxoDB was designed to provide a central point of access for all available T. gondii data, and a variety of data mining tools useful for the analysis of unfinished, un-annotated draft sequence during the early phases of the genome project. In later stages, as more and different types of data become available (microarray, proteomic, SNP, QTL, etc.) the database will provide an integrated data analysis platform facilitating user-defined queries across the different data types.
INTRODUCTION
Toxoplasma gondii is an intracellular apicomplexan parasite that infects humans, and virtually all warm-blooded organisms (1). T. gondii infection in healthy individuals is not normally associated with disease, but in association with pregnancy or immunosuppressive diseases and treatments, disease and death can occur (2,3). The genome is ∼80 Mb in size and consists of 11 chromosomes (4). T. gondii also serves as a model system for areas that are hard to investigate in the related malarial pathogen, Plasmodium falciparum (5,6).
It has been shown that much can be learned from EST (7) and unfinished draft sequence (8). The availability of draft sequence combined with databases and data mining tools (9) greatly facilitated research in the malaria and apicomplexan biology fields. The P. faliciparum genome has been sequenced to completion, but most eukaryotic genome projects, especially those in non-model organisms may not proceed all the way to completion. Numerous genomes, including the T. gondii genome will be sequenced via a random shotgun approach yielding 3–10X coverage. Organism-specific tools, such as gene finding algorithms, have yet to be developed or trained for most species. The challenge is to develop tools and strategies for handling unfinished draft sequence and maximize the amount of information that can be obtained from this increasingly common type of data, while avoiding the pitfalls inherent in interpreting unfinished datasets (redundancy in the data, sequencing and assembly errors, contaminating sequences, etc.).
DATA AND DATA MINING TOOLS
In building ToxoDB, we took advantage of existing database software and tools developed for the Allgenes (10,11) and PlasmoDB (9) databases. ToxoDB contains all publicly available genomic sequence data for two strains of T. gondii, RH and ME49-B7 and clustered EST assemblies from multiple strains representing all stages of the parasite lifecycle. As of the submission of this article, the database contains >70 million base pairs of nucleotide sequence including 36 333 contigs from genomic shotgun sequence, 10 239 BAC end-sequences, 773 BAC end-assemblies, and 6384 clustered EST sequences. These EST sequences, along with 6-frame translations of the genomic sequence (50 amino acids or greater), undoubtedly represent the majority of the coding potential in the T. gondii genome.
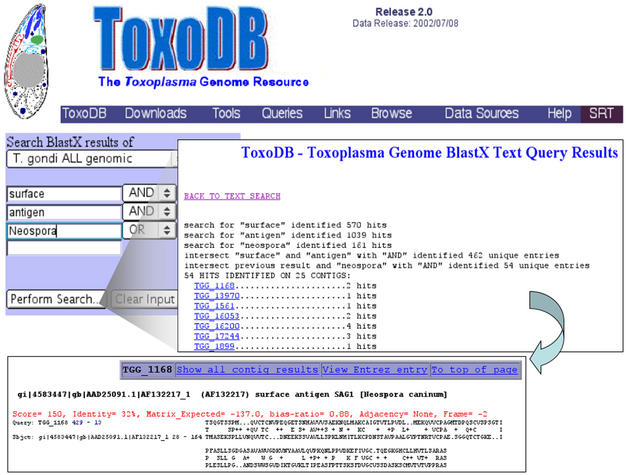
To facilitate gene discovery in T. gondii, we provide the user with several different tools. BLAST searches of six different databases are available (all genomic, all EST, all nucleotide data combined and the 6 frame ORF translations of each of these data sets). A protein motif finding tool is available for searching the above mentioned ORF amino acid databases for either known protein motifs, or user-defined motifs. A text search tool allows users to search the draft data using key words (Figure 1). The searchable text index was created by performing a BLASTX search of the NCBI non-redundant protein database with all of the draft sequences. The results of all BLAST searches with an E-value≤0.001 were stored and a text index of the words which appear in each BLAST hit definition line was created. In effect, users search for BLAST hits containing key words of interest and then they are able to view and retrieve the draft sequence that generated this hit. To provide an added measure of confidence, once a region of draft sequence is discovered, users can view a summary of all BLAST hits that were generated by the sequence and direct links to the NCBI GenBank are provided for each subject discovered by the search.
Figure 1.
Graphic illustration of the three phases of a text search. Users can select the database of BLAST results to be searched and provide up to 4 keywords joined by ‘AND’, ‘OR’ and ‘NOT’. In this case, the BLAST results for all T. gondii genomic sequences were searched for entries that contained the words, ‘surface’, ‘antigen’ and ‘Neospora’. The results summary page lists the search results for each keyword and the resulting join along with a hyperlink to the BLAST alignment containing the key words of interest. In this example, 54 hits were discovered that contained all three words. Hyperlinks in the BLAST alignment section permit users to view all BLAST hits generated by the particular draft contig (in this example TGG_1168) and connect directly to the entry for the ‘hit’ (AF132217) in the NCBI GenBank.
As users discover regions of interest, they have the ability to download the specific sequence region discovered in one of their searches, all sequences discovered, or any particular region they choose, (e.g. 1 kb upstream and downstream of a particular region) using quick links on the results page or specifically using the Sequence Retrieval Tool. A graphic tutorial on how to use all features of the database is located on the database home page.
DATA ACCESS AND DATA DEPOSITION
All sequence data contained on the site (nucleotide and ORF) are available for download subject to the data usage agreement (http://toxodb.org/). Sequences can be obtained individually, in FASTA format, as mentioned above, or in bulk, (e.g. all genomic sequence), in a variety of common file formats, (EMBL, FASTA and GenBank). A table of links to original data sources and an explanation of naming conventions are provided. Each sequence has a unique ID, a portion of which is traceable to the sequence source. Users are cautioned that due to the draft nature of the sequence, individual sequence IDs change for each genome assembly provided by the sequence source and hence in each database release. To facilitate the transition between releases, the previous version of the database will remain accessible via BLAST and the Sequence Retrieval Tool. A BLAST search of a new database release with draft sequences found in earlier searches will provide users with the new sequence ID.
ToxoDB can readily accept and process new sources of sequence data. As the genome project matures and additional types of genomic data are developed by the research community, ToxoDB is poised to accept and integrate this data with genomic sequence data. Any researcher interested in contributing data or suggestions on how the database can be improved to facilitate various types of research are urged to contact ToxoDB at help@toxodb.org.
FUTURE DIRECTIONS
As the genome project progresses and as more data types are generated, the focus of ToxoDB will shift from data mining of draft sequence to more sophisticated user-defined queries across integrated data types (e.g. find all predicted enzymes on chromosome 1b that have at least 1 trans-membrane domain and are up-regulated). The tools and infrastructure needed to capture and analyze and integrate a wide variety of data types are in place and will be similar to the types of queries and tools available for P. falciparum, at PlasmoDB.
Database enhancements scheduled for implementation in the near future include training of T. gondii gene finding algor-ithms, mapping of EST to genomic sequences, and stringent BLASTN searches of genomic sequence from other strains to the draft sequence from strain ME49-B7. Such mappings will help with gene finding, exon boundary determinations, and SNP discovery. Additionally, we will provide cross-species sequence comparisons among the Apicomplexa, particularly at the EST level.
Acknowledgments
ACKNOWLEDGEMENTS
Financial support for ToxoDB was provided by the National Institutes of Health. We wish to particularly thank the scientists who have collaborated with and contributed data to ToxoDB, and the software and database researchers at the University of Pennsylvania, who contributed greatly to the development and implementation of the software and infrastructure necessary to create organism-specific databases from generic software components. We also wish to thank the scientists, sequencing centers and funding agencies for agreeing to make the data available prior to publication. D.S.R. is the recipient of a Scholar Award in Molecular Parasitology from the Burroughs Wellcome Fund.
REFERENCES
- 1.Dubey J.P. (1997) Toxoplasma, Hammondia, Besnoita, Sarcocystis and other tissue cyst forming coccidia of man and animals. In Kreier,J.P. (ed.), Parasitic Protozoa III. Academic Press, New York, NY, pp. 101–237.
- 2.Remington J.S. and Desmonts,G. (1989) Toxoplasmosis. In Remington,J.S. and Klein,J.O. (eds), Infectious Diseases of the Fetus and Newborn Infant. W. B. Saunders, Philadelphia, PA, pp. 89–195.
- 3.Luft B.J. and Remington,J.S. (1992) Toxoplasmic encephalitis in AIDS patients. Clin. Infect. Dis., 15, 211–222. [DOI] [PubMed] [Google Scholar]
- 4.Sibley L.D. and Boothroyd,J.C. (1992) Construction of a molecular karyotype for Toxoplasma gondii. Mol. Biochem. Parasitol., 51, 291–300. [DOI] [PubMed] [Google Scholar]
- 5.Ajioka J.W. (1997) Analysis of apicomplexan parasites. Methods, 13, 79–80.9405191 [Google Scholar]
- 6.Roos D.S., Darling,J.A., Reynolds,M.G., Hager,K.M., Striepen,B. and Kissinger,J.C. (1999) Toxoplasma as a model parasite: apicomplexan biochemistry, cell biology, molecular genetics… and beyond. In Tschudi,C. and Pearce,E. (eds), Biology of Parasitism. Kluwer Press, Boston, MA, pp. 143–167.
- 7.Ajioka J.W., Boothroyd,J.C., Brunk,B.P., Hehl,A., Hillier,L., Manger,I.D., Marra,M., Overton,G.C., Roos,D.S., Wan,K.L., Waterston,R. and Sibley,L.D. (1998) Gene discovery by EST sequencing in Toxoplasma gondii reveals sequences restricted to the Apicomplexa. Genome Res., 8, 18–28. [DOI] [PubMed] [Google Scholar]
- 8.Janssen C.S., Barrett,P., Lawson,D., Quail,M.A., Harris,D., Bowman,S., Phillips,R.S. and Turner,C.M. (2001) Gene discovery in Plasmodium chabaudi by genome survey sequencing. Mol. Biochem. Parasitol., 113, 251–260. [DOI] [PubMed] [Google Scholar]
- 9.Bahl A., Brunk,B., Coppel,R.L., Crabtree,J., Diskin,S., Fraunholz,M.J., Grant,G.R., Gupta,D., Huestis,R.L., Kissinger,J.C., Labo,P., Li,L., McWeeney,S.K., Milgram,A.J., Roos,D.S., Schug,J. and Stoeckert,C.J.,Jr (2002) PlasmoDB: The Plasmodium Genome Resource. Nucleic Acids Res., 30, 87–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davidson S.B., Crabtree,J., Brunk,B., Schug,J., Tannen,V., Overton,G.C. and Stoeckert,C.J.,Jr (2001) K2/Kleisli and GUS: Experiments in integrated access to genomic data sources. IBM Systems J., 40, 512–531. [Google Scholar]
- 11.Stoeckert C.J. Jr, Pizarro,A., Manduchi,E., Gibson,M., Brunk,B., Crabtree,J., Schug,J., Shen-Orr,S. and Overton,G.C. (2001) A relational schema for array and non-array based gene expression data. Bioinformatics, 17, 300–308. [DOI] [PubMed] [Google Scholar]