Abstract
Two million plant ESTs, from 20 different plant species, and totalling more than one 1000 Mbp of DNA sequence, represents a formidable transcriptomic resource. Sputnik uses the potential of this sequence resource to fill some of the information gap in the un-sequenced plant genomes and to serve as the foundation for in silicio comparative plant genomics. The complexity of the individual EST collections has been reduced using optimised EST clustering techniques. Annotation of cluster sequences is performed by exploiting and transferring information from the comprehensive knowledgebase already produced for the completed model plant genome (Arabidopsis thaliana) and by performing additional state of-the-art sequence analyses relevant to today's plant biologist. Functional predictions, comparative analyses and associative annotations for 500 000 plant EST derived peptides make Sputnik (http://mips.gsf.de/proj/sputnik/) a valid platform for contemporary plant genomics.
INTRODUCTION
The publication of the Arabidopsis thaliana genome (1) and the draft sequences for two rice genomes (2,3) has provided a reference platform for plant genomics. The in-depth analysis of the known and predicted coding sequences from these plants has provided an invaluable resource on the gene-content for model plants and has allowed basic functional resolution of the plant genome.
The currently available plant genomes cover the basic gene repertoire needed for dicotyledonous (4) and monocotyledonous (5) plants. There is, however, a gene information deficit for the other plants that form model systems for e.g. root development in sugar beet, nitrogen-fixing root nodule formation in Lotus japonicus (6) or perhaps fruit development in avocado (7). There is also such an information deficit for researchers working on rapidly evolving and highly specific genes in other plant species. This information deficit is unlikely to be resolved by high throughput genome sequencing in the near-future. Genomic sequencing strategies are currently being undertaken in maize (http://mips.gsf.de/proj/maize/), Brassica oleracea (0.5–1×genome-sequencing coverage planned http://www.tigr.org/tdb/e2k1/bog1/) and Medicago truncatula (1×coverage in first phase http://www.genome.ou.edu/medicago.html) and various other projects are in the planning phases.
The remaining plant genomes remain inaccessible for the immediate future through either the technical issues associated with complete sequencing of the large and complex plant genomes or through the prohibitive costs associated with the sequencing and annotation of a complete genome (8).
Expressed-sequence tags (ESTs) (9) are typically short-length, low quality, contaminant enriched DNA sequences that have been produced by the sequencing of cDNA clones. EST sequences directly represent the transcriptome and avoid the typical genomic sequencing issues of highly-repetitive sequences. The preparation of the cDNA library in terms of tissues, stresses and developmental stages; and the subsequent normalisation of the library affects the overall sequence composition of EST collections. The expected information content is typically a largely redundant, highly heterogeneous, partially-overlapping collection of imperfect sequence reads—ideal starting material for bioinformatic analyses.
The computational processing of EST sequences has inherent problems with the data redundancy and quality. The redundancy within an EST collection, however, lends itself to computational methods for the clustering of the sequences to reduce the complexity of the dataset while concomitantly improving the overall quality of the data. A large EST collection when clustered to a stringent unigene collection has the potential to represent a significant proportion of the gene-coding portion of the genome (60.1% of A. thaliana genes contain a cognate EST-cluster match) (unpublished data).
While other research groups, most notably the TIGR Gene Indices (10) and the NCBI Unigenes (http://www.ncbi.nlm.nih.gov/UniGene/) have comprehensive collections of EST derived unigenes, our goal rather than to duplicate their efforts is to build, comprehensively annotate and maintain an up-to-date plant-specific EST derived sequence collection. We aim to exploit the available plant genomic data to perform heterologous associative annotation, and additionally to perform de novo sequence annotations using a variety of methods suited for the needs of the contemporary plant biologist and to support the gene modelling needs of the forthcoming plant genome projects.
ESTs AND ‘RECONSTRUCTOMICS’
Sputnik has been developed as a platform for ‘Reconstructomics’. The phrase reconstructomics has been coined to describe the application of genome-scale analyses and the transfer of annotation from complete plant genomes to partial genomes reconstructed from the available EST sequences. The extensive A. thaliana analysis database at MIPS (11) and the forthcoming rice genome database (Karlowski et al., this issue) lend themselves as a solid reference for the comparative annotation of sequences from the more ‘exotic’ EST derived plant genomes.
EST clustering and assembly is performed using the HarvESTer software (Biomax Informatics) on large public plant EST collections using parameters that encourage the clustering of paralagous sequences into different clusters. Protein sequences are predicted for all sequence assemblies on the basis of pre-calculated species-specific coding potentials. The EST, cluster and peptide sequences are collated in a relational database and are annotated in an automated manner using a suite of state-of-the-art bioinformatics resources.
The current challenge is to keep abreast with the rapidly growing public plant EST collections, to integrate, cluster and annotate the accrued sequence data, and to provide useful and biologist intuitive tools for the exploration and evaluation of the data. As a consequence of the enormous amounts of publicly released EST data and the needs of biologists within various projects, Sputnik is rapid evolving.
IMPLEMENTATION AND DATABASE STRUCTURE
Sputnik has been implemented as an EST, cluster and peptide management, annotation and data display pipeline. The application has been programmed as a collection of Python scripts that interact with a PostgreSQL relational database system. Sequences are typically imported from the EMBL format EST data available at the EBI (ftp://ftp.ebi.ac.uk). All native biological annotations from the sequence files are retained. These annotations contain such valuable information as the tissue used in the original cDNA library production, plant cultivar/variety information, and developmental stage information. Additionally, clone library information and any keywords or additional library descriptions are archived for subsequent searches. This infrastructure is additionally used on proprietary EST collections where available annotation on plant, cDNA library and tissue challenges are applied on an ad hoc basis.
Bioinformatic methods to be performed on sequences are defined within an XML file, thus allowing the simple inclusion of most bioinformatic methods that process sequence data, alignment data or the results from other bioinformatic methods. The analyses are processed in a heterogeneous distributed computational environment, and the raw results from the data are stored within the database structure and are related to the parent sequence accession. Post-processing is performed on the raw results from all analyses to allow for the fastest retrieval of all pertinent data and to simplify the interpretation and data display from the analyses.
Additional indexes are built upon the underlying raw sequence data to link multiple ESTs with a single unigene (either a cluster consensus or a singleton) and a single peptide prediction. This strict logical inheritance of sequence allows for the transfer of biological annotation (e.g. mRNA from a 3-week salt-stressed root cDNA library) and derived annotation from a single EST to a multi-member cluster to a peptide.
DATABASE CONTENTS
Currently, EST collections from all plant species with in excess of 10 000 public sequences have been integrated into Sputnik. Table 1 shows the plant EST collections available within Sputnik and basic statistics on the EST collections. The collection of species includes additional model species in terms of tuber development, fruit development, nodule association and other agronomically important plant species. In excess of 2 million ESTs have been analysed and resolved into ∼550 000 sequence clusters and singletons. The clustered sequences form the core basis for the annotation.
Table 1. Public plant EST collections currently available within the Sputnik databases.
| Organism | Common name | Number of ESTs | Total sequence (bp) | Number of cluster sequences |
|---|---|---|---|---|
| Glycine max | Soybean | 248 581 | 111 706 448 | 58 171 |
| Hordeum vulgare | Barley | 224 401 | 122 695 219 | 52 929 |
| Arabidopsis thaliana | Thale cress | 174 630 | 75 422 275 | 38 445 |
| Triticum aestivum | Wheat | 166 522 | 81 984 359 | 48 629 |
| Medicago truncatula | Barrel medic | 162 741 | 88 735 515 | 29 963 |
| Zea mays | Maize | 159 586 | 73 510 298 | 33 648 |
| Lycopersicon esculentum | Tomato | 148 350 | 74 566 527 | 30 012 |
| Chlamydomonas sp. | 112 609 | 62 638 867 | 30 632 | |
| Oryza sativa | Rice | 105 982 | 45 799 732 | 32 382 |
| Sorghum bicolor | Sorghum | 84 712 | 40 264 315 | 24 162 |
| Solanum tuberosum | Potato | 79 199 | 42 518 845 | 22 508 |
| Lactuca sativa | Lettuce | 76 592 | 44 989 782 | 26 602 |
| Physcomitrella patens | 67 738 | 32 606 312 | 18 803 | |
| Helianthus annuus | Sunflower | 64 733 | 31 026 007 | 13 739 |
| Pinus taeda | Pine | 49 293 | 20 336 883 | 17 414 |
| Lotus japonicus | Lotus | 31 566 | 11 870 293 | 11 102 |
| Gossypium arboreum | Cotton | 38 894 | 26 129 931 | 19 844 |
| Populus tremula×Populus tremuloides | 20 084 | 7 508 104 | 13 048 | |
| Mesembryanthemum crystallinum | Ice-plant | 17 190 | 10 574 948 | 9794 |
| Porphyra yezoensis | 10 354 | 4 777 904 | 3575 | |
| Secale cereale | Rye | 8122 | 3 772 940 | 5423 |
| Beta vulgaris | Sugar beet | 6029 | 3 132 192 | 3889 |
| Total | 2 057 908 | 1 016 567 696 | 544 714 |
For each parent species, the common plant name is shown along with information on the number of ESTs available, the total number of base-pairs sequenced and the number of sequence clusters that the ESTs have been resolved into.
The intrinsic sequence analyses performed within Sputnik reflect the needs of the modern plant molecular biologist and have been chosen within the context of collaborations within the GABI projects (http://www.gabi.de).
One of the key annotations to an EST derived sequence is the functional description. Sequences are functionally described by comparing the sequence against the MIPS catalogue of functionally described proteins (Funcat) (12). Sequences can be individually screened for candidate function on the basis of funcat reports. Whole sequence collections can be screened for sequences that have been assigned to a particular functional class using functional keywords and filtering the results by expectation values. Functional annotation is applied to both the cluster and peptide sequences, though in most cases the results are identical. In addition to functional annotation, structural annotation also lends itself to the elucidation of candidate roles for a particular peptide sequence. Structural annotation is performed using homology based approaches with both the SCOP domains database (13) and the PDB database of structurally resolved proteins (14). The results from these analyses allow for the selection of proteins that contain particular sequence folds or structures. All material is linked back to the source database. Domain annotation using the Interpro resource (15) again allows more extensible investigation of the domain content of a sequence. Interpro domains found are searchable by both keywords and Interpro descriptions. All Interpro domains are linked back to the additional information at the EBI.
The identification of molecular markers is a key component of many research projects. Polymorphic simple-sequence repeats (SSR) have been demonstrated to be present in both the gene-coding and the non gene-coding components of genomes (16,17). Cluster sequences have therefore been pre-screened for all putative perfect or imperfect SSRs up to the penta-nucleotide repeat size. Candidate SSRs can be selected as complete reports or as more specific repeats chosen on the basis of sequence composition, repeat size and overall ‘perfection’. SNPs are another fashionable sequence-marker that can be defined using bioinformatic algorithms (Rudd and Kota, manuscript in preparation). The cluster tiling path of multi-member clusters is screened for candidate SNP features, or residues at specific loci that differ from the consensus sequence. Correlation of the candidate SNPs with the EST sequence annotation on the plant variety/ecotype/subspecies can be used support or reject the hypothesis that this locus may represent a SNP, or may form only an allelic SNP. The putative SNPs identified are searchable on the basis of plant variety information and by SNP score produced by the underlying algorithm.
One of the appealing aspects of large EST collections produced from a variety of different tissues is the variety of information that can gleaned by correlating the relative numbers of sequences within specific clusters to either particular tissues, groups of tissues or even to specific challenges. This ‘poor-mans’ in silicio northern method has been implemented in Sputnik to allow investigation as to which sequences appear over-represented or under-represented within particular libraries and collections of libraries.
Comparative plant genomics is one of the underlying goals behind Sputnik. Sequence homology comparisons against the A. thaliana gene-set and the Arabidopsis genome scaffold can be used to assess the number of sequences that are shared with Arabidopsis, that are missing relative to Arabidopsis or that are absent in Arabidopsis. Such relationships can be simply investigated using a simple interface. All Arabidopsis homologies are linked back to the primary data within MAtDB so that the context of the sequences within a complete genome can be further understood. The comparative analyses have additionally been expanded into rice (MOsDB) and into the other reconstructed genomes within Sputnik.
QUERY INTERFACE
A basic internet based query interface provides access to Sputnik (Fig. 1). The main Sputnik page (http://mips.gsf.de/proj/sputnik/) provides links to all available Sputnik genomes. Selecting a species will display an introduction to the organism and the EST collection and provides access to the search methods implemented for each of the EST, cluster and peptide sequence levels. Selecting specific sequences is achieved by selecting a search method and supplying the required search parameters. Sequences can be searched for in terms of sequence accession, sequence length, cluster size or by annotated features. Annotated features include the functional and structural classifications, the molecular markers assessed and comparative analyses with respect to other genomes, or tissues and libraries within a single genome.
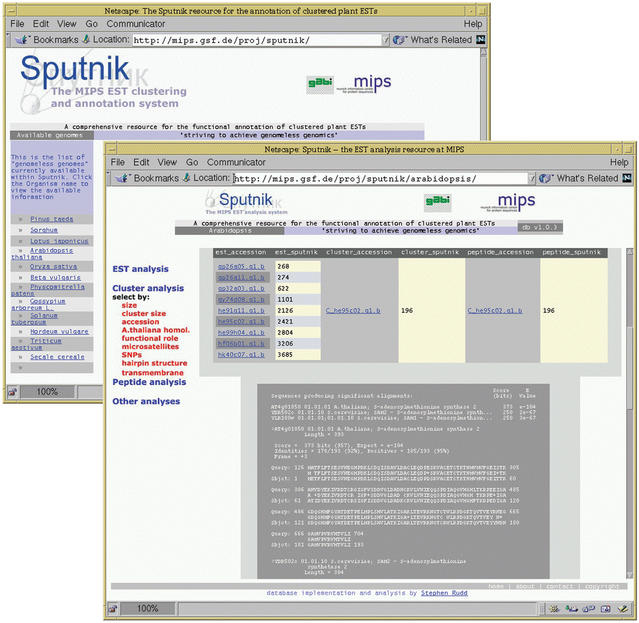
Figure 1.
Examples of Sputnik data views. (i) The main Sputnik page (underlying panel) provides information on the status and availability of the reconstructed plant genomes, and provides the links to the individual Sputnik database pages. Selecting a genome will display a window that provides access to the pre-calculated data and annotation (main panel). (ii) The project viewer provides links to search methods for the EST, cluster and peptide sequence layers. The available methods for the cluster layer are shown in red (left hand frame of main panel). (iii) For each sequence shown, links are provided to all accompanying ESTs, clusters or peptides (top of main panel), in this example links are provided from a single Arabidopsis cluster sequence to nine EST sequences and a single peptide. (iv) Data and annotation for each sequence is shown in the main frame, in the figure a functional report is shown for the Arabidopsis EST cluster, it corresponds to a S-adenosylmethionine synthase, 01.01.01 in the MIPS Funcat.
The sequence browser displays the primary sequence (cluster, EST or peptide) and provides links to the other related sequences (co-clustering ESTs, clusters or derived peptides). Additional links are provided to the data from the analysis and annotation methods. This data is pre-processed to provide hyperlinks back to the primary databases and to other extrinsic data sources.
DATA AVAILABILITY
The data within Sputnik are accessible via the WWW (http://mips.gsf.de/proj/sputnik/). The data is available free of charge in an unrestricted fashion to all academic users. Please contact the Sputnik administrator if you require the implementation of additional analyses, the inclusion of additional public or proprietary data or require any specific data not yet available from the database.
FUTURE DIRECTIONS
Sputnik is currently organized in a very project centric fashion. Future developments have been planned to adapt Sputnik to make it more suited to large-scale comparative plant genomics. We aim to include additional EST collections, to remain largely up to date with the ever-expanding plant EST collections and to integrate new sequence analyses and annotation methods. Currently, one of main problems with Sputnik is the vast amount of heterogeneous data available within the relational database structure that remains inaccessible to the casual web visitor. While the power of the relational databases is enormous, a simple to use interface to access all of the data in a simple manner is required. For this reason, we are in the process of indexing all Sputnik data within the BioRS application (Biomax informatics) that will allow extremely ambitious correlative analyses using multiple data sources within both single and multiple plant reconstructomes.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank the following collaborators for their invaluable contributions to the development of analyses and display interfaces; Markus Herz, Bernd Hackauf, Bernd Heidenreich, Georg Koch, Raja Kota, Silke Moehring, Uwe Hohmann, David Baulcombe, Eric Brenner and Shinhan Shiu. Grisha Kolesov and Axel Facius are involved in the mathematics and informatics of EST derived sequence markers. Andrea Hansen and Andreas Kaps (Biomax Informatics) have provided invaluable assistance with both the HarvESTer EST clustering application and in calculating EST clusters. Haruki Murakami provided inspiration with the naming of the database (ISBN: 0375411690). Sputnik is funded within the GABI project by the BMBF (0312270/4).
REFERENCES
- 1.The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 2.Goff S.A., Ricke,D., Lan,T.H., Presting,G., Wang,R., Dunn,M., Glazebrook,J., Sessions,A., Oeller,P., Varma,H. et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science, 296, 92–100. [DOI] [PubMed] [Google Scholar]
- 3.Yu J., Hu,S., Wang,J., Wong,G.K., Li,S., Liu,B., Deng,Y., Dai,L., Zhou,Y., Zhang,X. et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science, 296, 79–92. [DOI] [PubMed] [Google Scholar]
- 4.Allen K.D. (2002) Assaying gene content in Arabidopsis. Proc. Natl Acad. Sci. USA, 99, 9568–9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Livingstone K. and Rieseberg,L.H. (2002) Rice genomes: a grainy view of future evolutionary research. Curr. Biol., 12, R470–R471. [DOI] [PubMed] [Google Scholar]
- 6.Kawaguchi M., Imaizumi-Anraku,H., Koiwa,H., Niwa,S., Ikuta,A., Syono,K. and Akao,S. (2002) Root, root hair, and symbiotic mutants of the model legume Lotus japonicus. Mol. Plant Microbe Interact., 15, 17–26. [DOI] [PubMed] [Google Scholar]
- 7.Cowan AK., Cripps,RF., Richings,EW., and Taylor,NJ. (2001) Fruit size: towards an understanding of the metabolic control of fruit growth using avocado as a model system. Physiologia. Plantarum, 111, 127–136. [Google Scholar]
- 8.Adam D. (2000) Now for the hard ones. Nature, 408, 792–793. [DOI] [PubMed] [Google Scholar]
- 9.Adams M.D., Kelley,J.M., Gocayne,J.D., Dubnick,M., Polymeropoulos,M.H., Xiao,H., Merril,C.R., Wu,A., Olde,B., Moreno,R.F. et al. (1991) Complementary DNA sequencing: expressed sequence tags and human genome project. Science, 252, 1651–1656. [DOI] [PubMed] [Google Scholar]
- 10.Quackenbush J., Cho,J., Lee,D., Liang,F., Holt,I., Karamycheva,S., Parvizi,B., Pertea,G., Sultana,R. and White,J. (2001) The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res., 29, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoof H., Zaccaria,P., Gundlach,H., Lemcke,K., Rudd,S., Kolesov,G., Arnold,R., Mewes,H.W. and Mayer,K.F. (2002) MIPS Arabidopsis thaliana Database (MAtDB): an integrated biological knowledge resource based on the first complete plant genome. Nucleic Acids Res., 30, 91–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frishman D., Albermann,K., Hani,J., Heumann,K., Metanomski,A., Zollner,A. and Mewes,H.W. (2001) Functional and structural genomics using PEDANT. Bioinformatics, 17, 44–57. [DOI] [PubMed] [Google Scholar]
- 13.Lo Conte L., Brenner,S.E., Hubbard,T.J., Chothia,C. and Murzin,A.G. (2002) SCOP database in 2002: refinements accommodate structural genomics. Nucleic Acids Res., 30, 264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westbrook J., Feng,Z., Jain,S., Bhat,T.N., Thanki,N., Ravichandran,V., Gilliland,G.L., Bluhm,W., Weissig,H., Greer,D.S. et al. (2002) The Protein Data Bank: unifying the archive. Nucleic Acids Res., 30, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Apweiler R., Attwood,T.K., Bairoch,A., Bateman,A., Birney,E., Biswas,M., Bucher,P., Cerutti,L., Corpet,F., Croning,M.D. et al. (2000) InterPro—an integrated documentation resource for protein families, domains and functional sites. Bioinformatics, 16, 1145–1150. [DOI] [PubMed] [Google Scholar]
- 16.Tautz D. and Renz,M. (1984) Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res., 12, 4127–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kantety R.V., La Rota,M., Matthews,D.E. and Sorrells,M.E. (2002) Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol. Biol., 48, 501–510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data within Sputnik are accessible via the WWW (http://mips.gsf.de/proj/sputnik/). The data is available free of charge in an unrestricted fashion to all academic users. Please contact the Sputnik administrator if you require the implementation of additional analyses, the inclusion of additional public or proprietary data or require any specific data not yet available from the database.