Abstract
Arabidopsis thaliana is the most widely-studied plant today. The concerted efforts of over 11 000 researchers and 4000 organizations around the world are generating a rich diversity and quantity of information and materials. This information is made available through a comprehensive on-line resource called the Arabidopsis Information Resource (TAIR) (http://arabidopsis.org), which is accessible via commonly used web browsers and can be searched and downloaded in a number of ways. In the last two years, efforts have been focused on increasing data content and diversity, functionally annotating genes and gene products with controlled vocabularies, and improving data retrieval, analysis and visualization tools. New information include sequence polymorphisms including alleles, germplasms and phenotypes, Gene Ontology annotations, gene families, protein information, metabolic pathways, gene expression data from microarray experiments and seed and DNA stocks. New data visualization and analysis tools include SeqViewer, which interactively displays the genome from the whole chromosome down to 10 kb of nucleotide sequence and AraCyc, a metabolic pathway database and map tool that allows overlaying expression data onto the pathway diagrams. Finally, we have recently incorporated seed and DNA stock information from the Arabidopsis Biological Resource Center (ABRC) and implemented a shopping-cart style on-line ordering system.
INTRODUCTION
Arabidopsis thaliana is a small flowering plant that serves as a model organism for understanding the complex processes required for plant growth and development. The Arabidopsis Information Resource (TAIR's) goal is to provide an integrated, up-to-date view of Arabidopsis biology from genome to phenome from various sources of information ranging from personal communication with researchers to published literature. TAIR aims to facilitate interaction within a research community that collectively generates and refines a common body of knowledge (1–3). Since TAIRs inception, its usage has increased steadily from 20 000 web page visits per month in November 1999 to about 500 000 per month in August 2002 (http://arabidopsis.org/usage/). Likewise, the content of the database has increased dramatically, largely due to incorporation of data from large-scale genomics projects and stock centers. Current, detailed statistics of TAIR database content are available on-line (http://arabidopsis.org/jsp/tairjsp/pubDbStats.jsp). TAIR continues to evolve and expand to include new data types and analysis tools. It is committed to improving data content by continuous curation and data analysis facilitated by collaborations with and feedback from the research community. Detailed information about the TAIR project, including the full proposal, data sources, projects under development and documentation of the software and curation procedures are available on-line (http://arabidopsis.org/about/) and in a comprehensive review (4).
UPDATED LOCUS INFORMATION
The genome sequence increased the number of Arabidopsis genes identified about 20-fold. There are currently 28 974 sequenced, physical loci (defined as a non-overlapping region of the genome corresponding to at least one transcription unit) and 549 loci defined genetically. All physical loci follow a standard nomenclature that is based on the genomic location and is synchronized among collaborating databases (TAIR, MIPS' MATDB, TIGR's ATH1 and NCBI's RefSeq). More information about locus and gene nomenclature is found on-line (http://arabidopsis.org/info/guidelines.html). Genes that correspond to the same physical or genetic locus but have different structural or functional annotations or produce different transcribed products are stored as separate gene models and are associated to the corresponding locus. There are currently 35 184 gene models in TAIR. Organization and structural annotation of these genes on the genome are essential and continual processes, aided by improved analysis methods and incorporation of experimental data supporting the gene models. As a consequence of the large-scale re-annotation effort by The Institute for Genome Research (TIGR, http://www.tigr.org/tdb/e2k1/ath1/), some of the original loci have been merged or split. Therefore, some locus names have been made obsolete and new locus names have been created. The history of the locus names and the current usage can be searched and downloaded (http://arabidopsis.org/tools/bulk/locushistory/). One of our major efforts has been identifying and grouping gene models with different annotations, including names, arising from many sources such as TIGR, MIPS, GenBank and the literature. There are now over 66 000 aliases including ORF names, protein names, full names and 4800 gene symbols. Aliases have been designated based upon sequence match or published allelism data.
GENOME ANNOTATION AND VISUALIZATION
The use of structured, shared vocabularies to describe the roles of genes and products will facilitate identifying similarities among diverse organisms and revealing the extent or absence of knowledge gathered. TAIR is a member of the Gene Ontology Consortium (http://www.geneontology.org), which has developed widely-adopted structured vocabularies to describe gene products (5). Our main concentration has been modifying structures and adding terms to better accommodate annotation of plant genes. Both TAIR and TIGR are actively annotating Arabidopsis gene products with GO terms using information from the literature, sequence similarity and other computational methods. Currently, TAIR has annotated 13 041 unique gene models with GO terms for gene product function, process and cellular component. We have also developed controlled vocabularies to describe Arabidopsis anatomy and stages of development (ftp://ftp.arabidopsis.org/home/tair/Ontologies/) and are working to extend these ontologies to include other plant species, in conjunction with several plant genome databases [Gramene (http://www.gramene.org/), MaizeDB (http://www.agron.missouri.edu/) and International Rice Research Institute (http://www.irri.org/)] under the auspices of the Plant Ontology Consortium.
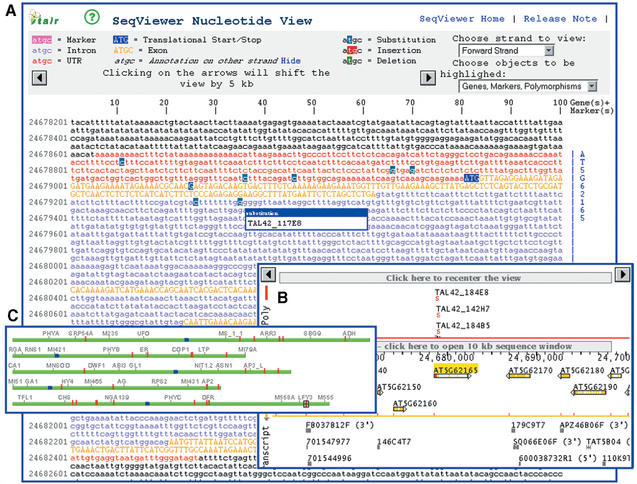
TAIRs SeqViewer (http://arabidopsis.org/servlets/sv) allows viewing of the Arabidopsis genome decorated with genes, genetic markers, clones, polymorphisms and transcripts. Researchers can interactively choose to display or hide various objects and select different zoom levels from a whole chromosome down to a 10 kb nucleotide view. Search options include text (e.g. clone, marker, transcript, polymorphism or gene names, up to 250 names at a time) or short nucleotide sequences (no more than 4 sequences of 15–150 nucleotides each). Multiple hits are displayed on the genome, in a close-up graphical view or in a nucleotide window (Fig. 1). Matching entities can be viewed by clicking on the genome to open a close-up graphical view or by clicking on a match summary to open a table-format view. Several close-up views can be opened in the same or different windows to allow users to compare regions on the same or different chromosomes.
Figure 1.
TAIRs SeqViewer. (A) Nucleotide view showing 10 kb of sequence with genes indicated as colored letters (UTRs in red, exons in gold, introns in purple) with translational starts and stops highlighted in blue and mutated nucleotides highlighted in gray. (B) Close up view showing a 40 kb region surrounding the gene visible in A, with mutations and polymorphisms shown at top (poly band), genes shown in center (gene band) and EST matches shown at bottom (transcript band). (C) Genome view showing the five Arabidopsis chromosomes in green with the positions of genes matching a name search indicated as vertical red lines.
A nucleotide window displays 10 kb of any region of the genome, with all the selected sequenced entities highlighted on the actual nucleotide sequence (Fig. 1). The view can be scrolled up or down 5 kb at a time, and allows users to view genes on either one or both strands with UTRs, start and stop codons, exons and introns highlighted in different colors. EST and full-length cDNA matches are displayed as dashed lines. Genetic markers and different types of polymorphisms such as insertions, deletions, substitutions and compound (a combination of either substitution/insertion or substitution/deletion) are highlighted in different colors. Ends of clones are also be precisely located in the nucleotide window. Sequence from this page can be copied and pasted into other applications with the gene annotation preserved as uppercase (coding regions) and lowercase (introns, UTRs and intergenic regions). A complementary tool, MapViewer (http://arabidopsis.org/servlets/mapper), can be used to compare Arabidopsis sequence, physical and genetic maps (1).
METABOLIC PATHWAYS, ENZYMES, REACTIONS AND COMPOUNDS
Biochemical pathways provide another way to group gene products and related data, including compounds, co-factors, reactions, enzymes, enzyme complexes and subcell enzyme locations. TAIR has relied on the Pathway Tools software (6) to build an Arabidopsis specific pathway database called AraCyc (http://arabidopsis.org/tools/aracyc). It currently features over 1800 enzymes and 1108 enzymatic reactions involved in 174 pathways containing 822 different compounds. AraCyc pathways were generated computationally, then edited manually to correct mistakes, remove non-plant pathways and add missing plant pathways. To date, more than twenty plant-specific pathways, including carotenoid, brassinosteroid and gibberellin biosyntheses have been added from the literature.
The main query page for AraCyc provides links to descriptions of the data set, an overview map, and the results of the initial computational build of the database. The overview map gives a bird's-eye view of all pathways in the database, which can be overlaid with expression patterns of genes. Gene expression values (or any other attributes of genes in numeric forms) can be highlighted on the lines representing reactions by uploading a file containing gene names and attribute (e.g. expression, sequence similarity score, etc.) values. The tool also supports text-based queries to find pathways, reactions, proteins, genes, or compounds, as well as browsing a classification hierarchy of pathways, Enzyme Commission (http://www.chem.qmul.ac.uk/iubmb/enzyme/) reactions, compounds or genes. Pathways are displayed graphically at different levels of detail down to the molecular structures of all compounds and co-factors. All of the reactions, compounds, genes, proteins and cofactors on pathways are hyperlinked to detail pages. Genes are also hyperlinked to TAIRs locus detail pages.
MICROARRAYS, POLYMORPHISMS AND OTHER FUNCTIONAL GENOMICS EFFORTS
TAIR is in the process of incorporating expression data from microarray experiments from users and external databases. Besides the quantitative values of expression of the arrayed elements, the expression data class includes detailed information about the experimental design and the source of the RNA used for hybridization, as well as the data analysis and protocols employed. The first set of data being incorporated represents the bulk of experiments performed by the Arabidopsis Functional Genomics Consortium (AFGC) (http://afgc.stanford.edu/afgc_html/site2.htm), which consists of about 560 hybridizations.
Information about sequence polymorphisms can be utilized for genetic marker development, whole genome analysis and comparative analysis. TAIR database contains over 12 000 genetic markers and 100 000 polymorphisms submitted by the public research community. In addition, TAIR provides access to the collection of 56 670 predicted Arabidopsis single-nucleotide polymorphisms (SNP) and small insertions/deletions (INDELs) generated by Cereon Genomics (http://arabidopsis.org/Cereon/index.html). Cereon has also released ∼95 Mb of sequence from the Ler strain, which is comprised of 81 306 sequence entries from a single-pass shotgun sequencing (7). The Cereon data are accessible as HTML pages, downloadable tab-delimited text files, or via BLAST (for Ler sequence) only to registered users from non-profit institutions, universities and colleges.
‘The 2010 Project’ was proposed by the Arabidopsis community as a follow-up to the AGI genome sequencing, with the goal of understanding the function of all Arabidopsis genes by the year 2010 (8). As a result of this initiative, functional genomic projects designed to identify the molecular function of all Arabidopsis genes have been initiated all over the world. The role of TAIR is to provide an infrastructure to facilitate dissemination of the information generated from these projects. Information about currently funded functional genomic projects can be found at http://arabidopsis.org/info/2010_projects/. Anyone can use this site to search for the genes included as subjects of a funded research proposal, as well as obtain information about the individual projects and researchers. This information is also available in the database from the Locus and Community detail pages.
DOWNLOADING DATA SETS
Data can be downloaded from TAIR in several ways. Upon obtaining results from the text-based queries, check boxes allow researchers to download a tab-delimited file of selected results. For larger datasets relating to gene and protein information (e.g. protein characteristics, GO annotations, gene-related DNA and protein sequences, and microarray elements) can be queried and downloaded in bulk by pasting in or uploading a set of locus names (http://arabidopsis.org/tools/bulk/).
In addition to downloading from the database, TAIR provides and updates a variety of large data files in tab-delimited and FASTA formats from the FTP site (ftp://ftp.arabidopsis.org/home/tair/). Updates of most files are performed on a regular basis. The collection includes all sequence data sets used by the sequence analysis programs (Arabidopsis CDS, transcript, protein, intron, exon, intergenic, locus upstream or downstream sequences, all higher plant sequences, Brassica sequences), the complete Arabidopsis genome sequences (nuclear, chloroplast and mitochondrial), all physical and genetic mapping data, a variety of gene and protein data files, microarray data files, structured vocabularies and structural and functional gene annotation files. Specialized, large data sets currently not on the FTP site can be obtained by contacting curators at curator@arabidopsis.org.
BIOLOGICAL MATERIALS AND STOCK ORDERING
TAIR recently integrated the information from ABRC (http://arabidopsis.org/abrc/) and implemented an ordering system formerly managed by Arabidopsis Information Management System [AIMS, (9)]. TAIR can now be used to obtain information about the DNA and seed stocks available at the stock center, to place orders on-line and to view the history of orders for any stock. The sequenced BAC, EST, and full-length cDNA clones, in conjunction with the mapping resources of the stock center, provide a powerful set of resources for positional gene cloning, complementation, expression and protein characterization (9). Pooled genomic DNA obtained from pooled transgene insertion lines is also available for PCR screening to identify insertions in a gene of interest, complementing the ‘knockout’ services of the Arabidopsis Knockout Facility (AKF) (http://www.biotech.wisc.edu/Arabidopsis/).
ABRC maintains and distributes seeds from public and private sources. Mapping resources include mapped mutant lines, multiple marker lines, four populations of recombinant inbred lines, trisomic lines and a population organized by tetrads. Over 140 000 transposon and T-DNA populations with random insertions throughout the genome have been produced, largely by the SALK institute (http://signal.salk.edu/). Currently, there are 50 256 insertion flank sequences available, about half of which are associated to loci. These and other insertion flank sequences are mapped on the genome and included as a data set for BLAST and the other sequence analysis programs offered at TAIR. Large numbers of characterized lines with transpositions to random locations are also available, together with lines transformed with specific transgenes and molecular tags, transposon parental stocks and natural variants collected in the wild from around the world.
IMPLEMENTATION
TAIR uses an object-oriented approach to data representation and software architecture (4). The underlying database is implemented in a relational database management system (Sybase version 11.9.2). More detailed information about the database schemas and software documentation can be found on-line (http://arabidopsis.org/about/).
DATA SUBMISSION AND USER FEEDBACK
We encourage researchers to participate in making TAIR more usable and useful by submitting data, correcting errors and providing feedback on the usability of TAIR. We are particularly interested in gene annotation corrections and updates, gene family information, genetic marker information and functional genomics characterization information. More information on how to submit these data can be found on-line (http://arabidopsis.org/info/data_submission.html). In addition, researchers can enter their notes or annotations on every data detail page by clicking on the ‘ADD MY NOTES’ button. Currently, only community registration and update functions are fully supported by interactive, electronic submission process. Our main goal in the upcoming year is to develop easier and more rigorous ways of allowing direct data submission by the researchers.
DOCUMENTATION AND USER SUPPORT
Currently, four types of user support are provided: (1) Email communication. Each page generated at TAIR contains email addresses for contacting: curator@arabidopsis.org for general questions, comments, and bug reports, dnastock@arabidopsis.org for questions regarding DNA stocks and orders and seedstock@arabidopsis.org for questions regarding seed stocks and orders; (2) Help buttons on the headers of each page point to elaborated help documentation on using different tools at TAIR; (3) Names of column headers and data descriptors are hyperlinked to an appropriate section of the help documentation and (4) A searchable ‘Frequently Asked Questions’ (http://arabidopsis.org/help/) allow users to explore similar questions that had been answered previously.
HYPERLINKING TO TAIR DATA PAGES
TAIR detail pages can be hyperlinked by using the base URL, http://arabidopsis.org/servlets/TairObject, and providing either TAIR accession or a combination of name and type of the data class. All major data classes in TAIR have a unique accession, which is a combination of the class type followed by a colon and the identifier (e.g. Gene: 1944475). Relevant TAIR accessions are displayed on all detail pages. In addition, a list of TAIR accessions can be downloaded from the FTP directory as well as from text-based search results. More detailed information about hyperlinking to TAIR is available on-line (http://arabidopsis.org/about/linktotair.html).
CITING TAIR
The following citation format is suggested when referring to specific datasets or information from TAIR: The Arabidopsis Information Resource (TAIR), Carnegie Institution, Stanford, CA and National Center for Genome Resources, Santa Fe, NM [http://arabidopsis.org, (month, year)]. Please use this paper when referencing TAIR.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the TAIR users and the Arabidopsis research community for their continuing support, feedback, and particularly for sharing their data and expertise. TAIR is supported by NSF grants DBI-9978564 and DBI-0091471. GO development and annotation is supported in part by NIH/NHGRI grant HG-02273. This is Carnegie Institution of Washington Department of Plant Biology Publication 1550.
REFERENCES
- 1.Huala E., Dickerman,A.W., Garcia-Hernandez,M., Weems,D., Reiser,L., LaFond,F., Hanley,D., Kiphart,D., Zhuang,M., Huang,W. et al. (2001) The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res., 29, 102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reiser L., Mueller,L.A. and Rhee,S.Y. (2002) Surviving in a sea of data: a survey of plant genome data resources and issues in building data management systems. Plant Mol. Biol., 48, 59–74. [PubMed] [Google Scholar]
- 3.Rhee S.Y. (2000) Bioinformatic resources, challenges, and opportunities using Arabidopsis as a model organism in a post-genomic era. Plant Physiol., 124, 1460–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Hernandez M., Berardini,T., Chen,G., Crist,D., Doyle,A., Huala,E., Knee,E., Miller,N., Mueller,L.A., Mundodi,S. et al. (2002) TAIR: a resource for integrated Arabidopsis data. Func. Integ. Genomics, 2, 239–253. [DOI] [PubMed] [Google Scholar]
- 5.The Gene Ontology Consortium (2001) Creating the Gene Ontology Resource: design and implementation. Genome Res., 11, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karp P.D., Riley,M., Paley,S.M. and Pellegrini-Toole,A. (2002) The MetaCyc Database. Nucleic Acids Res., 30, 59–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jander G., Norris,S.R., Rounsley,S.D., Bush,D.F., Levin,I.M. and Last,R.L. (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol., 129, 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chory J., Ecker,J.R., Briggs,S., Caboche,M., Coruzzi,G.M., Cook,D., Dangl,J., Grant,S., Guerinot,M.L., Henikoff,S. et al. (2000) National Science Foundation-Sponsored Workshop Report: ‘The 2010 Project’. Functional genomics and the virtual plant: a blueprint for understanding how plants are built and how to improve them. Plant Physiol., 123, 423–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scholl R., May,S. and Ware,D. (2000) Seed and molecular resources for Arabidopsis. Plant Physiol., 124, 1477–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]