Abstract
Organellar Genome Retrieval (OGRe) is a relational database of complete mitochondrial genome sequences for over 250 Metazoan species. OGRe provides a resource for the comparative analysis of mitochondrial genomes at several levels. At the sequence level, OGRe allows the retrieval of any selected set of mitochondrial genes from any selected set of species. Species are classified using a taxonomic system that allows easy selection of related groups of species. Sequence alignments are also available for some species. At the level of individual nucleotides, the system contains information on base frequencies and codon usage frequencies that can be compared between organisms. At the level of whole genomes, OGRe provides several ways of visualizing information on gene order. Diagrams illustrating the genome arrangement can be generated for any selected set of species automatically from the information in the database. Searches can be done based on gene arrangement to find sets of species that have the same order as one another. Diagrams for pairwise comparison of species can be produced that show the positions of break-points in the gene order and use colour to highlight the sections of the genome that have moved. OGRe is available from http://www.bioinf.man.ac.uk/ogre.
INTRODUCTION
Mitochondrial genome sequences are widely used in the evolutionary studies of many different groups of organisms. They range in size from just under 6 kb in Plasmodium falciparum and Plasmodium reichenowi to over 360 kb in some plants such as Beta vulgaris and Aribidopsis thaliana. The 69 kb mitochondrial genome of Reclimonas americana is the least derived and most gene rich discovered to date, containing 94 genes and bearing a clear relationship to its eubacterial ancestor (1). Most metazoan mitochondrial genomes are much smaller (∼16 kb) and highly derived (2); they code for a reduced set of 13 proteins, 22 tRNAs and two rRNAs.
Although the gene content of metazoan mitochondria is highly consistent, the order of those genes varies substantially between different taxa. At the time of writing, we observe 14 unique gene orders amongst the set of 177 available complete vertebrate mitochondrial genomes and 50 unique gene orders amongst the 75 invertebrate genomes. This reflects the fact that there are many closely related vertebrate species having identical gene orders, whilst for the invertebrate phyla, where the species sampling is less dense, almost all species differ from one another in gene order. As the number of invertebrate genomes available increases, we expect to observe a rapid increase in the number of unique gene orders. Several mechanisms are envisaged for the rearrangement of mitochondrial genomes including inversions, translocations, duplications and deletions (3). Gene order information can also be used for phylogenetic purposes (3–5). Various algorithms, including inversion distances, breakpoints and related edit distances, having been developed for measuring distances between gene orders and subsequently reconstructing phylogenetic trees (6–11).
In addition to gene orders, mitochondrial gene sequences have proved extremely popular in molecular phylogenetics. They have been used for reconstructing phylogenies at various different levels—from intraspecies (12) to the divergence of large taxa (13,14). The conserved sets of single copy orthologous genes found in complete mitochondrial genomes greatly facilitate the reconstruction of combined gene phylogenies (15).
Variation in the frequencies of nucleotide bases can bias the results of phylogenetic methods (16,17). Base frequencies on mitochondrial genomes differ quite substantially between species, sometimes even between those that are closely related. In addition, it has been shown that base frequencies can vary between the two DNA strands and sometimes from one part of the genome to another (18,19). Mitochondria provide a closed system within which to study variations in base frequency and therefore codon usage. The frequencies of synonymous codons in mitochondrial genes are closely dependent on the base frequencies, suggesting that the asymmetry of mutational rates between the four bases is one of the most important factors in determining codon usage patterns (20). It is also possible, however, that selective effects such as increasing efficiency of translation or avoiding certain unfavourable DNA motifs may play a role.
Gene order, combined gene phylogeny and codon usage within mitochondria are related progressive areas of research. Large-scale sequencing programs such as those on protists and fungi (1,21), invertebrates (22), fish (23,24) and mammals (25,26) mean that there are now over 300 complete mitochondrial genome sequences available in the public databases. We have designed a database, Organellar Genome Retrieval, (OGRe) to extract the complete sequence data and provide new resources based around gene order information and codon usage data. Here we describe OGRe and the facilities that it provides.
THE CONTENTS OF OGRe
OGRe is a relational database designed to store and provide easy access to organelle genome sequences and related information. At the time of writing, OGRe contains the mitochondrial genomes of 252 metazoan species. We have limited ourselves to metazoa in this first release of OGRe because this is the group for which most genomes are available, and the level of evolutionary divergence is appropriate both for phylogenetics and for gene order comparisons. We note, however, that OGRe has been designed to cope with all organellar sequences including those that contain introns (the only species currently in the database with an intron-containing gene is Metridium senile). Our policy is only to include species for which a finished genome record is available in the NCBI list of complete organelle genomes (http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/euk_o.html). This results in the inclusion of a few species whose sequences are referred to as ‘complete’ by NCBI, but which in fact have one or two missing genes.
The data in OGRe are checked by hand and we correct many annotation errors in the original NCBI GenBank data set. We have found annotation errors to be particularly frequent in tRNA genes. In particular, genes were sometimes not labelled or were placed on the wrong strand. Missing genes and strand errors are evident when sequences are compared in alignments. There were also cases where we judged the start and stop positions of genes to be misplaced—again this is evident when looking at alignments of large numbers of species.
OGRe differs from the other databases that cater for organelle genomes in both operation and additional content. OGRe provides a taxonomic hierarchy specifically designed to facilitate the downloading of single or multiple sequences, display of codon usage information and diagrams of genomes at any level of classification in one go. In addition, OGRe provides complete RNA gene alignments for many species as well as tools to compare whole genomes and search for species sharing gene orders.
SELECTING SPECIES
Retrieving data from OGRe is organism driven. This means that the user must first select the organisms they are interested in and then select the data they wish to retrieve. There are three methods of choosing organisms; from an alphabetical list of all the organisms within the database, by selecting individual organisms or groups of organisms using the Taxonomy Browser or by searching for a specific organism by partial Latin or common names. Although some of these features are similar to methods of organism selection in the other mitochondrial databases—GOBASE (27) and AMmtDB (28)—we provide a system that we believe allows faster and simpler access to the required data.
From the alphabetical list of genomes, any desired combination of species may be selected and raw information for the genomes of individual organisms may be retrieved, including specific annotations, MEDLINE references and a link back to the original GenBank record.
The Taxonomy Browser Allows the user to select individuals or groups of organisms using the hierarchical taxonomic system stored in the database. This is displayed as a collapsible tree in which taxa may be expanded to give access to further levels of subclassification until finally reaching individual species. The level of detail of the sub-groups depends on the number of available genomes within the groups. For phyla with few complete genomes (e.g. Annelida, Nematoda etc.) the phylum is the smallest subdivision. Extra detail will be added to these parts of the classification as and when necessary. For Chordates and Arthropods the present taxonomy extends to a more detailed level. The taxonomy that we use is based upon the NCBI's classification (http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html), although we have included far fewer levels of subdivision. It is possible to select all species contained in any taxon in one go. For example, one may select either all Chordates, or all Mammals, or all Rodents, depending on the level of detail required.
Sequences may also be selected from searches using either latin name or common name. Having chosen species using any of the methods above, the user may elect either to display sequences from those genomes, to view the genomes, or to display codon usage information using the options at the foot of the species selection pages.
RETRIEVING SEQUENCES
Choosing to display sequences allows the user to select genes from those present in the selected organisms. The names of the genes are the same as those used in GenBank. Having selected one or more genes the appropriate sequences may be viewed or downloaded in FASTA format. FASTA format has been chosen as it is recognized by almost all sequence analysis programs (e.g. for sequence alignment or phylogenetics). When downloading protein sequences, there is an option to display them as either nucleotides or amino acids. For amino acid sequences, OGRe does the translation using the appropriate genetic code for the species concerned. We note that this is considerably more convenient than downloading the sequences from GenBank, which would require a separate download of each sequence.
As well as being able to download raw sequences from the database, we provide sequence alignments of mitochondrial genes, again in FASTA format, for use with phylogenetic studies. As alignments are completed, they will be made available from the OGRe ‘gene alignments’ page. We provide structure-based alignments of RNA genes that indicate, using bracket notation, the conserved secondary structure of the genes on the top line of the alignment file. In preparing our RNA alignments we have taken account of established secondary structure patterns where appropriate for tRNAs (29,30) and rRNAs (31). These files can be used with our RNA-based phylogenetic software PHASE (see http://www.bioinf.man.ac.uk/resources/). PHASE uses our specific evolutionary models for compensatory mutations in the helical regions of RNA structures (32,33). Alignments of the mammalian RNA genes we have used in testing PHASE (15) are currently available.
CODON USAGE TABLES
Two codon usage tables may be displayed for each selected species—one for each strand. It is known that base frequencies, particularly those at the third codon position, are not equal on the two strands (20). This is thought to arise from mutations occurring during the DNA replication process, when the two strands are separate. For this reason, codon frequencies in genes on the two strands are significantly different. Although codon usage information for mitochondrial sequences is available from the Japanese codon usage database (34) (http://www.kazusa.or.jp/codon), the strand effect is not accounted for. This particular system also includes multiple copies of some genes for organisms that have been sequenced more than once, which means that codon counts cannot be used for statistical tests such as chi-square tests. In contrast, codon counts in OGRe include exactly one copy of each gene.
VIEWING AND COMPARING WHOLE GENOMES
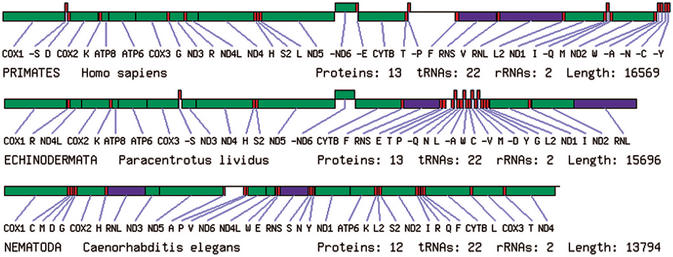
We provide the facility to dynamically generate a diagram of the genome for each selected species. The genomes are represented as linear rather than circular in order to save space and to allow several genomes to be seen on one page. This feature is not provided by existing databases. OGRe produces the diagrams using the gene position information stored in the database. Boxes are used to illustrate the size and position of each gene and whether it is on the forward or reverse strand. Colours are used to distinguish between protein, rRNA and tRNA genes. We adopt the convention used by Boore (Mitochondrial gene arrangement source guide, http://www.jgi.doe.gov/Mitochondrial_Genomics.html) that the COX1 gene is placed first in the sequence. Figure 1 shows the mitochondrial genome diagrams of human (Homo sapiens), a sea urchin (Paracentrotus lividus) and a nematode (Caenorhabditis elegans), as examples. The latter genome lacks the ATP8 gene and has all genes on one strand.
Figure 1.
An example of three genome diagrams produced by the OGRe genome viewer.
OGRe can be searched for species that share gene orders. Using the gene order information system, any single species may be selected. The database returns a schematic diagram of the gene order of that species together with a list of other organisms that share the same gene order. Some analyses of genome rearrangements ignore tRNA genes because they move more frequently than the other genes the option therefore exists to search the database with or without taking tRNAs into account.
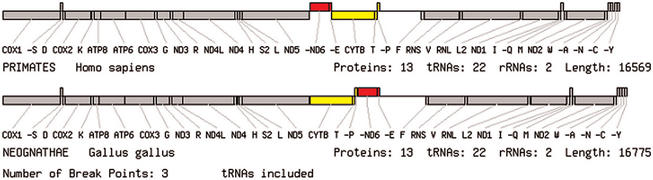
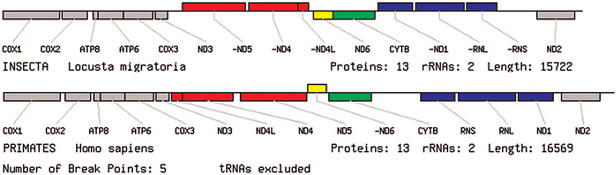
In order to facilitate pairwise comparisons between genomes, we have provided a specialized version of the genome viewer. The differences between any two genomes selected by the user from the ‘genome comparison’ page are shown by diagrams with the same layout as the genome viewer, but a different colour scheme is used to illustrate genome rearrangements. The two genomes are divided into uninterrupted blocks (where the gene order is the same in both genomes) separated by break points. Break points are the ends of regions that have undergone inversion or translocation (8). Genes in different blocks are coloured differently, whilst matching blocks on the two genomes are given the same colour. The number of break points between the two genomes is also shown in the output. Figure 2 shows a comparison of human and chicken (Gallus gallus) genomes. In this case, two blocks of genes have swapped positions, which could have occurred because of a translocation or by a duplication followed by deletions of one copy of each gene. Again, the option exists to ignore tRNAs when calculating the break points and colouring the diagram. Figure 3 shows a comparison of human and locust that excludes tRNAs. In this case, there are 5 break points and the pattern of inversions of the major genes is easy to see, whereas there are 21 break points between these species when tRNAs are included.
Figure 2.
An example of a pairwise genome comparison produced by OGRe where tRNA genes are included. The blocks of genes coloured red and yellow have exchanged positions between human and chicken but all genes remain on the same strand.
Figure 3.
An example of a pairwise genome comparison produced by OGRe where tRNA genes are excluded. There are apparently three separate inversions of blocks of genes between human and locust (shown in red, yellow and blue). The grey and green gene blocks are unchanged.
THE FUTURE OF OGRe
OGRe is updated with complete metazoan mitochondrial genomes as they are released by the NCBI and checked by us for errors. We would welcome input from readers interested in the mitochondrial genomes of plants, fungi, protists as well as chloroplast genomes who might wish to help in the expansion of our system. Expansion to these additional species will involve additional work on gene identification of a much wider range of genes.
Having established the basis of the OGRe database in this release, we will build on it by adding additional novel features in future, including more explicit details of RNA secondary structures, a wider range of sequence alignments, and phylogenetic trees produced from mitochondrial sequences.
Acknowledgments
ACKNOWLEDGEMENTS
D.J., A.P.G. and C.H. are all supported by the Biotechnology and Biological Sciences Research Council of the UK. P.G.H. is supported by the Canada Research Chairs organization. We also wish to thank the following students on the University of Manchester MSc Bioinformatics course who have contributed to earlier versions of the Taxonomy Browser and genome viewer: Sian Dibben, David Gerrard, Rahbeia Jamil, Nathan Johnson and Paul Pennington. We are grateful to Jane Mabey for advice on using the PostgreSQL database management system.
REFERENCES
- 1.Lang F., Seif,E., Gray,M.W., O'Kelly,C.J. and Burger,G. (1999) A comparative genomics approach to the evolution of eukaryotes and their mitochondria. J. Eukaryot. Microbiol., 46, 320–326. [DOI] [PubMed] [Google Scholar]
- 2.Gray M.W. (1999) Evolution of organellar genomes. Curr. Opin. Genet. Dev., 9, 678–687. [DOI] [PubMed] [Google Scholar]
- 3.Boore J.L. and Brown,W.M. (1998) Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr. Opin. Genet. Dev., 8, 668–674. [DOI] [PubMed] [Google Scholar]
- 4.Stechmann A. and Schlegel,M. (1999) Analysis of the complete mitochondrial genome of the brachiopod Terebratulina retusa places the Brachiopoda with the protostomes. Proc. R. Soc. Lond. Biol. Sci., 266, 2043–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roehrdanz R.L., Degrugillier,M.E. and Black,W.C. (2002) Novel rearrangements of arthropod mitochondrial DNA detected with long PCR: applications to arthropod phylogeny and evolution. Mol. Biol. Evol., 19, 841–849. [DOI] [PubMed] [Google Scholar]
- 6.Bafna V. and Pevzner,P.A. (1996) Genome rearrangements and sorting by reversals. SIAM J. Comput., 25, 272–289. [Google Scholar]
- 7.Bafna V. and Pevzner,P.A. (1998) Sorting by transpositions. SIAM J. Discrete Math., 11, 224–240. [Google Scholar]
- 8.Blanchette M., Kunisawa,T. and Sankoff,D. (1999) Gene order break point evidence in animal mitochondrial phylogeny. J. Mol. Evol., 49, 193–207. [DOI] [PubMed] [Google Scholar]
- 9.Sankoff D. and Blanchette,M. (1999) Phylogenetic invariants for genome rearrangements. J. Comp. Biol., 6, 431–445. [DOI] [PubMed] [Google Scholar]
- 10.Larget B., Simon,D.L. and Kadane,J.B. (2002) Bayesian phylogenetic inference from animal mitochondrial genome arrangements. J. R. Stat. Soc. Ser. B, in press. [Google Scholar]
- 11.Bourque G. and Pevzner,P.A. (2002) Genome–scale evolution: reconstructing gene orders in ancestral species. Genome Res., 12, 26–36. [PMC free article] [PubMed] [Google Scholar]
- 12.Ingman M., Kaessmann,H., Pääbo,S. and Gyllensten,U. (2000) Mitochondrial genome variation and the origin of modern humans. Nature, 408, 708–713. [DOI] [PubMed] [Google Scholar]
- 13.Waddell P.J., Cao,Y., Hauf,J. and Hasegawa,M. (1999) Using novel phylogenetic methods to evaluate mammalian mtDNA. Syst. Biol., 48, 31–53. [DOI] [PubMed] [Google Scholar]
- 14.Mindell D.P., Sorenson,M.D., Dimcheff,D.E., Hasegawa,M., Ast,J.C. and Yuri,T. (1999) Interordinal relationships of birds and other reptiles based on whole mitochondrial genomes. Syst. Biol., 48, 138–152. [DOI] [PubMed] [Google Scholar]
- 15.Jow H., Hudelot,C., Rattray,M. and Higgs,P.G. (2002) Bayesian phylogenetics using an RNA substitution model applied to early mammalian evolution. Mol. Biol. Evol., 19, 1591–1601. [DOI] [PubMed] [Google Scholar]
- 16.Foster P.G. and Hickey,D.A. (1999) Composition bias may affect both DNA-based and protein-based phylogenetic reconstructions. J. Mol. Evol., 48, 284–290. [DOI] [PubMed] [Google Scholar]
- 17.Schmitz J., Ohme,M. and Zischler,H. (2002) The complete mitochondrial sequence of Tarsius bancanus: evidence for extensive nucleotide compositional plasticity of primate mitochondrial DNA. Mol. Biol. Evol., 19, 544–553. [DOI] [PubMed] [Google Scholar]
- 18.Saccone C., De Giorgi,C., Gissi,C., Pesole,G. and Reyes,A. (1999) Evolutionary genomics in metazoa: the mitochondrial DNA as a model system. Gene, 238, 195–209. [DOI] [PubMed] [Google Scholar]
- 19.Saccone C., Gissi,C., Reyes,A., Larizza,A., Sbisa,E. and Pesole,G. (2002) Mitochondrial DNA in metazoa: degree of freedom in a frozen event. Gene, 286, 3–12. [DOI] [PubMed] [Google Scholar]
- 20.Reyes A., Gissi,C., Pesole,G. and Saccone,C. (1998) Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol. Biol. Evol., 15, 957–966. [DOI] [PubMed] [Google Scholar]
- 21.Gray M.W., Lang,B.F., Cedergren,R., Golding,B.G., Lemieux,C., Sankoff,D., Turmel,M., Brossard,N., Delage,E., Littlejohn,T.G. et al. (1998) Genome structure and gene content in protist mitochondrial DNAs. Nucleic Acids Res., 26, 865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boore J.L. (1999) Animal mitochondrial genomes. Nucleic Acids Res., 27, 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue J.G., Miya,M., Tsukamoto,K. and Nishida,M. (2001) A mitogenomic perspective on the basal teleostean phylogeny: resolving higher-level relationships with longer DNA sequences. Mol. Phylogenet. Evol., 20, 275–285. [DOI] [PubMed] [Google Scholar]
- 24.Miya M., Kawaguchi,A. and Nishida,M. (2001) Mitogenomic exploration of higher teleostean phylogenies: A case study of moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol. Biol. Evol., 18, 1993–2009. [DOI] [PubMed] [Google Scholar]
- 25.Cao Y., Fujiwara,M., Nikaido,M., Okada,N. and Hasegawa,M. (2000) Interordinal relatationships and timescale of eutherian evolution as inferred from mitochondrial genome data. Gene, 259, 149–158. [DOI] [PubMed] [Google Scholar]
- 26.Arnason U., Adegoke,J.A., Bodin,K., Born,E.W., Esa,Y.B., Gullberg,A., Nilsson,M., Short,R.V., Xu,X. and Janke,A. (2002) Mammalian mitogenomic relationships and the root of the eutherian tree. Proc. Natl Acad. Sci. USA, 99, 8151–8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimko N., Liu,L., Lang,B.F. and Burger,G. (2001) GOBASE, the organelle genome database. Nucleic Acids Res., 29, 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lanave C., Licciulli,F., De Robertis,M., Marolla,A. and Attimonelli,M. (2002) Update of AMmtDB: a database of multi-aligned Metazoa mitochondrial DNA sequences. Nucleic Acids Res., 30, 174–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinberg S., Leclerc,F. and Cedergren,R. (1997) Structural rules and conformational compensations in the tRNA L-form. J. Mol. Biol., 266, 269–282. [DOI] [PubMed] [Google Scholar]
- 30.Helm M., Brulé,H., Friede,D., Giegé,R., Pütz,J. and Florentz,C. (2000) Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA, 6, 1356–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cannone J.J., Subramanian,S., Schnare,M.N., Collett,J.R., D'Souza,L.M., Du,Y., Feng,B., Lin,N., Madabusi,L.V., Muller,K.M. et al. (2002) The Comparative RNA Web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics, 3, http://www.biomedcentral.com/1471-2105/3/2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higgs P.G. (2000) RNA secondary structure: physical and computational aspects. Q. Rev. Biophys., 33, 199–253. [DOI] [PubMed] [Google Scholar]
- 33.Savill N.J., Hoyle,D.C. and Higgs,P.G. (2001) RNA sequence evolution with secondary structure constraints: comparison of substitution rate models using maximum likelihood methods. Genetics, 157, 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura Y., Gojobori,T. and Ikemura,T. (2000) Codon usage tabulated from the international DNA sequence databases: status for the year 2000. Nucleic Acids Res., 28, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]