Abstract
PlasmoDB (http://PlasmoDB.org) is the official database of the Plasmodium falciparum genome sequencing consortium. This resource incorporates the recently completed P. falciparum genome sequence and annotation, as well as draft sequence and annotation emerging from other Plasmodium sequencing projects. PlasmoDB currently houses information from five parasite species and provides tools for intra- and inter-species comparisons. Sequence information is integrated with other genomic-scale data emerging from the Plasmodium research community, including gene expression analysis from EST, SAGE and microarray projects and proteomics studies. The relational schema used to build PlasmoDB, GUS (Genomics Unified Schema) employs a highly structured format to accommodate the diverse data types generated by sequence and expression projects. A variety of tools allow researchers to formulate complex, biologically-based, queries of the database. A stand-alone version of the database is also available on CD-ROM (P. falciparum GenePlot), facilitating access to the data in situations where internet access is difficult (e.g. by malaria researchers working in the field). The goal of PlasmoDB is to facilitate utilization of the vast quantities of genomic-scale data produced by the global malaria research community. The software used to develop PlasmoDB has been used to create a second Apicomplexan parasite genome database, ToxoDB (http://ToxoDB.org).
CONTENTS OF THE CURRENT RELEASE
Data: complete sequence for Plasmodium falciparum, annotation and functional genomics datasets
Integrated efforts at The Institute for Genome Research, the Sanger Institute, and the Stanford University Genome Technology Center have produced an effectively complete genome sequence for P. falciparum strain 3D7 (1). The finished sequence is featured in PlasmoDB (2) version 4.0, released October 2002 (3). New sequence data is also available for P. yoelii and other Plasmodium species.
Also new to PlasmoDB 4.0 are large-scale datasets derived from proteomics analysis of several life cycle stages (4), expression profiling results from throughout the intraerythrocytic cycle (oligonucleotide-based microarrays in both glass-slides and Affymetrix formats), and single nucleotide polymorphism (SNP) analysis for P. falciparum strains HB3, DD2, D10, 7G8 and 3D7 (5). Protein and RNA data can be used to identify genes expressed in different life stages. Expression data can be analyzed with clustering software, e.g. XCluster (http://genome-www.stanford.edu/~sherlock/cluster.html), or used to look for differentially expressed genes using PaGE (6). Genes containing these SNPs can be retrieved and assessed for non-synonymous amino acid changes.
The urgent need to identify potential drug and vaccine targets has driven the P. falciparum genome sequencing project from its inception (7). In addition to DNA sequence data and analyses of predicted genes and proteins, Gene Ontology (GO) assignments have been provided by the sequencing centers and others in the malaria research community, and this curated annotation greatly facilitates drug target discovery. To assist in the identification of potential vaccine targets, annotated genes have been scored for potential T-cell epitopes using the SYFPEITHI method (8).
Tools and queries
The PlasmoDB 4.0 web interface incorporates several improvements over previous releases. A new version of the ‘gene display’ page makes more extensive use of graphical elements to present a concise single-page summary for each annotated gene in the database. This summary page allows users to quickly examine both the genomic arrangement of a gene (intron/exon structure, placement and identity of neighboring genes, etc) and its likely function, based on graphical summaries of predicted protein features (signal peptides, protein motifs, transmembrane domains, etc) and pre-computed database search results. Other pages linked to the gene present specialized views of relevant data, such as predicted mRNA and protein sequences, detailed protein motif predictions and microarray expression results. PlasmoDB now provides direct links to external data sources and sites, including the Malaria Parasite Metabolic Pathways database (http://sites.huji.ac.il/malaria/) and the Malaria Research and Reference Reagent Resource Centre, MR4 (9).
In addition to supporting new queries, release 4.0 improves the ease with which queries run against the relational database GUS can be combined with other data analysis tools, and with previously run queries. For example, one can now query for all genes in a subtelomeric region of chromosome 4 that contain a user-defined protein motif and with an additional click or two, these results can be downloaded as a FASTA-formatted list. The first part of this query uses a (new) SQL query against the relational database, while the second part uses the ‘Amino Acid Motif Search’ tool; these tools are combined through the ‘query history’ feature of the web interface.
PlasmoDB 4.0 enables several queries that exploit the new data types present in this release (Table 1). For example, one can quickly retrieve all predicted genes for which at least two lines of experimental evidence (e.g. mass spectrometric analysis of proteins and oligonucleotide-based microarray data) suggest expression in merozoites. Polymorphism queries permit identification of (for example) all genes with non-synonymous amino acid changes in the HB3 strain relative to the 3D7 strain. Queries on pre-computed BLAST results have been extended to incorporate taxonomic information, supporting (for example) queries for P. falciparum genes that are closely conserved in at least one other Plasmodium species but have no apparent homolog in the human genome.
Table 1. Summary of new data types and related new queries present in PlasmoDB 4.0.
| New data | Sample queries |
|---|---|
| Completed P. falciparum genome sequence and first-pass annotation. Additional genomic sequence for several other Plasmodium species. New protein feature analyses. | Find P. falciparum genes with (or without) homologs in other Plasmodium species? Which proteins containing putative T-cell epitopes? |
| SNPs for P. falciparum strains 3D7,HB3,DD2, D10, 7G8 (chromosome 3 only; whole genome to follow). | Identify genes with known sequence polymorphisms? Which SNPs yield non-synonymous substitutions? |
| Additional SAGE data. Additional gene expression data from cDNA and oligonucleotide-based glass slide microarray. Expression data from Affymetrix chips. | Find genes that are differentially expressed using the PaGE algorithm? Use Xcluster to group expression profiles? |
| Proteomic mass spectrometry data from multiple P. falciparum lifecycle stages. | Find genes for which both microarray and proteomics evidence indicates expression in a specified lifecycle stage? |
Plasmodium falciparum GenePlot
GenePlot was designed to provide researchers around the world with access to the genome sequence and annotations for the malaria parasite P. falciparum. Access is available online, or through a stand-alone CD-ROM that does not require high-speed internet connectivity (10). The re-written and greatly enhanced, release of GenePlot contains complete P. falciparum genome sequence, all annotations provided by the sequencing centers, gene predictions from three applications (trained on P. falciparum data), DNA sequence repeats, pre-computed TBLASTX analysis of the entire genome sequence, BLASTP similarities of all predicted genes and protein feature predictions for all predicted and annotated genes.
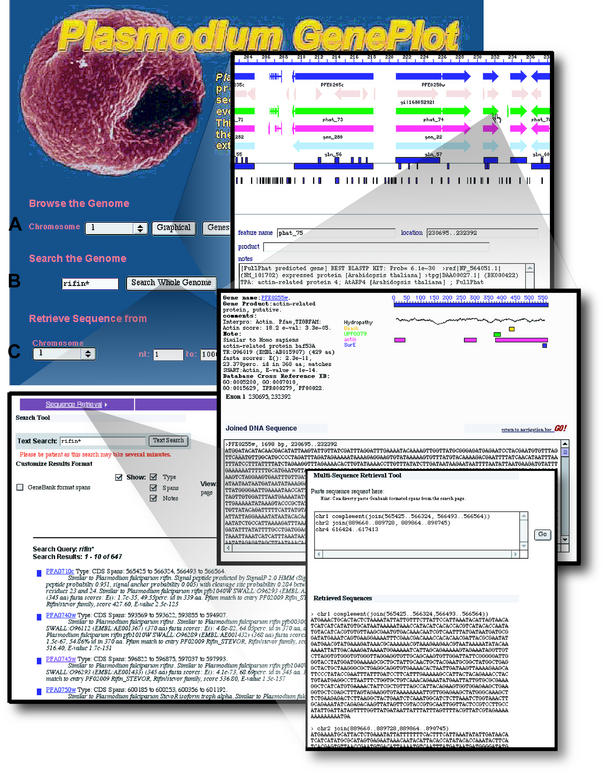
As illustrated in Figure 1, a graphical interface permits browsing the genome and genome annotations, including predicted and annotated protein features. Search capabilities allow compound text-based queries of curated gene/protein annotations, automated analyses and the results from pre-computed comparisons with GenBank/EMBL and other relevant databases. Annotated and predicted gene sequences can be selectively retrieved using the genome search interface and sequences can be retrieved in multiple formats from many different contexts. A tutorial on GenePlot usage is also provided.
Figure 1.
Composite of screen displays demonstrating features of the CD, Plasmodium GenePlot. The GenePlot home page provides access to three main tools. A graphical genome browser (A) highlights curated gene models (dark blue arrows), BLAST hits (shades of red reflect P-values), gene/coding potential predictions based on several different algorithms (PHAT, green arrows; GeneFinder, pink arrows; GlimmerM, light blue arrows; Hexamer, blue boxes), and tandem sequence repeats. Graphical views present chromosome data for individual genes and features via mouse-over; clicking on any one of these pulls up a page providing further information and evidence on all features associated with the gene. The search tool (B) performs complex searches of all genomic features and creates reports in a variety of different formats. Nucleotide sequences can be obtained from either the home page (C) or individual gene pages, using a multiple sequence retrieval tool. Some of the sequence manipulation tools available are shown in the background.
P. falciparum GenePlot can be accessed directly or downloaded from the PlasmoDB web site. The CD-ROM version is also available free of charge (along with other materials of interest to malaria researchers) from helpcd@plasmodb.org or via the Malaria Research and Reference Reagent Resource Center (MR4); malaria@atcc.org. Email requests should include ‘Nature malaria CD-ROM’ in the subject line and a full postal address in the body of the message.
FUTURE PLANS
With the official release of the finished P. falciparum genome sequence, a large influx of new data is anticipated from functional genomics studies, including expression profiling, proteomics, population genetics and other projects. Redesign of the display and query infrastructure will allow users to set and save preferences defining how DNA sequences, genes, proteins and expression data should be viewed and downloaded. Users will also be able to store queries for use in future sessions and share lists of genes with other interested PlasmoDB users. The underlying GUS architecture (11; http://www.gusdb.org) has already been exploited to develop a database for the related parasite Toxoplasma gondii (12), and other organism-specific applications can be envisaged.
Acknowledgments
ACKNOWLEDGEMENTS
Financial support for PlasmoDB was provided by the Burroughs Wellcome Fund, and the database was developed using computational infrastructure from the Liniac project at the University of Pennsylvania Genomics Institute. We thank the numerous researchers who have collaborated with and contributed to PlasmoDB by depositing both published and unpublished data, by making software available and by making useful suggestions on how to improve this community resource. We wish to thank the scientists and funding agencies comprising the international Malaria Genome Project for making sequence data from the genome of P. falciparum (3D7) public prior to publication of the completed sequence. The Sanger Institute provided sequence for chromosomes 1, 3–9 and 13, with financial support from the Wellcome Trust. A consortium involving The Institute for Genome Research and the Naval Medical Research Center sequenced chromosomes 2, 10, 11 and 14, with support from NIAID/NIH, the Burroughs Wellcome Fund and the Department of Defense. The Stanford Genome Technology Center sequenced chromosome 12, with support from the Burroughs Wellcome Fund.
REFERENCES
- 1.Gardner M.J., Hall,N., Fung,E., White,O., Berriman,M., Hyman,R.W., Carlton,J.M., Pain,A., Nelson,K.E., Bowman,S. et al. (2002) The genome sequence of the human malaria parasite Plasmodium falciparum. Nature, 419, 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahl A., Brunk,B., Coppel,R.L., Crabtree,J., Diskin,S.J., Fraunholz,M.J., Grant,G.R., Gupta,D., Huestis,R.L., Kissinger,J.C. et al. (2002) Plasmo-DB: The Plasmodium genome resource. Nucleic Acids Res., 30, 87–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kissinger J.C., Brunk,B.P., Crabtree,J., Fraunholz,M.J., Gajria,B., Milgram,A.J., Pearson,D.S., Schug,J., Bahl,A., Diskin,S.J. et al. (2002) PlasmoDB: The Plasmodium genome resource. Nature, 419, 490–492. [DOI] [PubMed] [Google Scholar]
- 4.Florens L., Washburn,M.P., Raine,J.D., Anthony,R.M., Grainger,M., Haynes,J.D., Moch,J.K., Muster,N., Sacci,J.B., Tabb,D.L. et al. (2002) A proteomic view of Plasmodium falciparum life cycle. Nature, 419, 520–526. [DOI] [PubMed] [Google Scholar]
- 5.Mu J., Duan,J., Makova,K.D., Joy,D.A., Huynh,C.Q., Branch,O.H., Li,W.H. and Su,X.Z. (2002) Chromosome-wide SNPs reveal an ancient origin for Plasmodium falciparum. Nature, 418, 323–326. [DOI] [PubMed] [Google Scholar]
- 6.Manduchi E., Grant,G.R., McKenzie,S.E., Overton,G.C., Surrey,S. and Stoeckert,C.J.,Jr (2000) Generation of patterns from gene expression data by assigning confidence to differentially expressed genes. Bioinformatics, 16, 685–698. [DOI] [PubMed] [Google Scholar]
- 7.Fletcher C. (1998) The Plasmodium falciparum genome project. Parasitol. Today, 14, 342–344. [DOI] [PubMed] [Google Scholar]
- 8.Donnes P. and Elofsson,A. (2002) Prediction of MHC class I binding peptides using SVMHC. BMC Bioinformatics, 3, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Y. and Rogers,M.J. (2002) Shared knowledge can combat malaria. Nature, 419, 15. [DOI] [PubMed] [Google Scholar]
- 10.Milgram A.J., Gajria,B., Kissinger,J.C., Pearson,D.S. and Roos,D.S. (2002) Plasmodium falciparum GenePlot (CD-ROM). Nature, 419, in press. [Google Scholar]
- 11.Davidson S., Crabtree,J., Brunk,B.P., Schug,J., Tannen,V., Overton,G.C. and Stoeckert,C.J.,Jr (2001) K2/Klesli and GUS: Experiments in integrated access to genomic data sources. IBM Systems J., 40, 512–531. [Google Scholar]
- 12.Kissinger J.C., Gajria,B., Li,L., Paulsen,I. and Roos,D.S. (2003) ToxoDB: Accessing the Toxoplasma gondii genome. Nucleic Acids Res., 31, 234–236. [DOI] [PMC free article] [PubMed] [Google Scholar]