Abstract
Zea mays DataBase (ZmDB) seeks to provide a comprehensive view of maize (corn) genetics by linking genomic sequence data with gene expression analysis and phenotypes of mutant plants. ZmDB originated in 1999 as the Web portal for a large project of maize gene discovery, sequencing and phenotypic analysis using a transposon tagging strategy and expressed sequence tag (EST) sequencing. Recently, ZmDB has broadened its scope to include all public maize ESTs, genome survey sequences (GSSs), and protein sequences. More than 170 000 ESTs are currently clustered into ∼20 000 contigs and about an equal number of apparent singlets. These clusters are continuously updated and annotated with respect to potential encoded protein products. More than 100 000 GSSs are similarly assembled and annotated by spliced alignment with EST and protein sequences. The ZmDB interface provides quick access to analytical tools for further sequence analysis. Every sequence record is linked to several display options and similarity search tools, including services for multiple sequence alignment, protein domain determination and spliced alignment. Furthermore, ZmDB provides web-based ordering of materials generated in the project, including ESTs, ordered collections of genomic sequences tagged with the RescueMu transposon and microarrays of amplified ESTs. ZmDB can be accessed at http://zmdb.iastate.edu/.
INTRODUCTION
Maize is a major world crop and an important model organism for addressing fundamental questions in monocotyledonous plants, which include rice, wheat and all the other cereals that together comprise a group of crops of unsurpassed historical, economic and human nutritional importance. Zea mays DataBase (ZmDB; http://www.zmdb.iastate.edu) is a public database that serves modern maize genetics by linking genomic sequence data with gene expression data and phenotypic information on mutant plants. As part of the on-going Maize Gene Discovery, Sequencing and Phenotypic Analysis Project (MGDP; 1), ZmDB serves as the Web portal to project-generated data, including expressed sequence tag (EST) sequences, data from microarrays consisting of representative EST sets designed to cover all of the identified maize genes, genomic survey sequences (GSSs) derived from the flanking regions of RescueMu transposon insertions in the maize genome (2,3), and phenotypic descriptions of mutant plants recovered from RescueMu-tagged populations. When available, the transposon-derived GSSs are linked to phenotypic descriptions of their likely source plants. Conversely, the associated maize phenotype database can be queried for particular phenotype descriptions, which may then be traced to the potential causative transposon insertions. More recently, ZmDB has broadened its scope to include all public maize ESTs, GSSs, and protein sequences. Gene-level comparisons across different plant species are facilitated through integration of ZmDB into the larger context of a new database effort, PlantGDB (http://www.plantgdb.org/).
DATABASE COMPONENTS
SequenceDB
At an estimated 2500 Mb, the maize genome is comparable in size to the human genome. Its highly repetitive nature (resulting from nested retrotransposon insertions that likely comprise 75% of the genome) has made a direct sequencing strategy for gene discovery impractical (4). Instead, the maize research community thus far has favored two alternative approaches to gene identification. One approach relies on EST sequencing, a strategy that has proven tremendously successful for gene discovery in other organisms, either as a stand-alone approach or in conjunction with whole genome sequencing. The second approach consists of generating maize GSSs that are enriched in genic regions by a variety of methods, including transposon tagging, hypomethylation filters and selection for long open reading frames (reviewed in 5;6). Currently, most of the contributions to the maize GSS data derive from the flanking regions of RescueMu transposon insertions sequenced by the MGDP.
ZmDB staff regularly imports maize EST and GSS sequences from the GenBank dbEST and dbGSS divisions, respectively. This procedure has been adopted to preserve the unique GenBank accession numbers. All imported sequences enter the ZmDB annotation pipeline. The ESTs are processed with ZmDBAssembler (http://www.zmdb.iastate.edu/zmdb/EST/assembly.html), a portable Perl script integrating several external programs, including BLAST (7) for initial clustering of the ESTs and CAP3 (8) for the ultimate assembly and consensus sequence determination. The assembly produces tentative unique genes (TUGs), which comprise tentative unique contigs (TUCs—clusters with two or more member ESTs) and tentative unique singlets (TUSs—ESTs that are not significantly similar to any other ESTs). The current collection of over 170 000 maize ESTs has been assembled into ∼20 000 TUCs and an about equal number of TUSs.
Presently, maize GSSs consist of over 100 000 entries of average length 400 bp. Each GSS is screened for matching maize ESTs and potential gene products by the spliced alignment software GeneSeqer using maize-specific splice site prediction parameters (9). RescueMu transposons are recovered for use as sequencing templates after digestion of total maize DNA with two restriction enzymes (http://zmdb.iastate.edu/zmdb/library-plate/GridGprep.html); the resulting right and left sequences flanking each RescueMu element are trimmed when the restriction site(s) is encountered; consequently, any particular RescueMu clone can produce up to four genomic DNA sequence fragments. A second feature of the strategy is that somatic RescueMu insertions should be sequenced only once while heritable insertions are likely to be recovered several times. We use the GSSAssembler program (Q. Dong, unpublished) to remove the redundancy in the RescueMu-derived GSSs and to derive a GSS-based maize gene index. GSSAssembler incorporates similar principles as the ZmDBAssembler, but uses the vmatch algorithm (10) instead of BLAST for more accurate and faster clustering. Presently, over 15 000 GSS contigs have been assembled from RescueMu-derived GSSs.
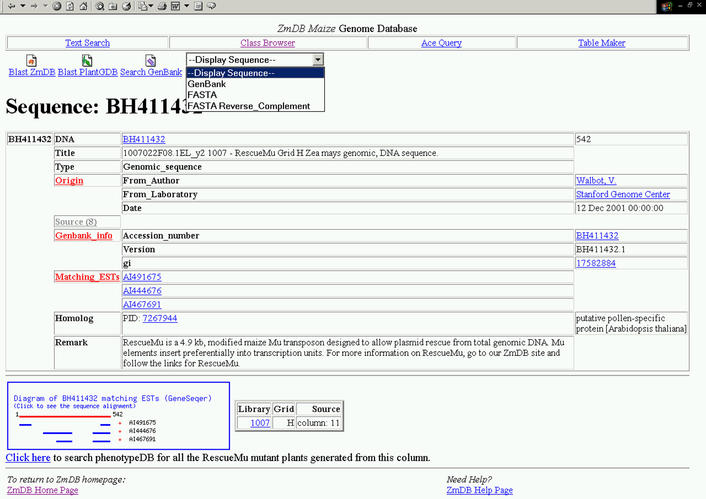
Functional annotation of the ESTs, GSSs and their assembled consensus sequences is largely automated. BLAST searches are performed against the most current non-redundant GenBank databases. BLAST output is processed with the previously described MuSeqBox program (11), and up to three highly significant matches are reported in the ZmDB records. The protein sequences are also annotated with respect to sequence motifs. The design philosophy of ZmDB is to go beyond providing a static picture of these analyses by providing dynamic functionality at the site. Users can quickly re-analyze the data with different tools, parameter settings, or external data. Thus, ZmDB has a ‘work-bench’ look and feel to its interface. Figure 1 shows a typical GSS entry display, in which tools are integrated for analysis. For example, clicking on the ‘Blast PlantGDB’ icon brings the GSS sequence to our PlantGDB search page. From there, users cannot only search against maize sequences but also perform cross-species comparisons against any combinations of other major plant EST sets.
Figure 1.
Display of GSS entries at ZmDB. The screenshot represents a typical display of the GSS records at ZmDB. The central panel provides basic annotation for the sequence, including key GenBank features and links to matching EST records. Source annotation can be viewed when selected. Sequence similarity search tools (BLAST) are linked via the icons on top (selection pastes the sequence into the query screen of the respective tool). The diagram in the lower left panel displays a schematic of the spliced alignment of significantly matching ESTs derived with the GeneSeqer program. The actual alignments and alignment scores can be viewed by clicking on the diagram. Blue lines delineate putative exons. In this example, AI444676 and AI467691 indicate two exons, but EST AI491675 corresponds to an alternative transcript. The small table to the right of the diagram links to the source DNA sample, and the link below facilitates a search of the PhenotypeDB for possible source plants.
PhenotypeDB
This component database describes and catalogues mutant maize plants generated by MGDP. Plants containing RescueMu as well as native Mu elements were planted in grids of 48 rows by 48 columns each. These plants are self-pollinated and the progeny evaluated. Phenotype information of each selfed individual is recorded at three developmental stages: seed and ear, seedlings in the greenhouse and field-grown plants through adult reproduction. Screening has been completed for 23 000 seed and ear stages, 7000 seedling families and 7000 adult plants. Mutant phenotypes are described by a customary controlled vocabulary that is extensively documented at the web site. Images of plants with novel characteristics are available at the web site.
There are currently three inter-connected query interfaces to serve different needs: (i) phenotype lists, which contain text descriptions of all phenotype terms and their corresponding abbreviations stored in the database. This search can find specific kinds of mutations, e.g. plants that are male sterile; (ii) mutant browser, which was designed to locate broad classes of mutations, e.g. plants with color deficiencies and (iii) location search, which displays phenotype information about all plants in specific locations, e.g. all plants from Grid ‘G’ Row ‘2’. In addition to the phenotype data, tools are also provided for researchers to facilitate tracing the cause of the observed mutant traits. For example, bi-directional links were established between traits and RescueMu insertion sites when available. On detailed phenotype mutant display pages, researchers can query the ZmDB sequence collections for all the RescueMu GSSs generated from the same row or column in which the mutant plant is located. On RescueMu GSS display pages, researchers can query the PhenotypeDB for mutant plants that grew in the same row or column as where the sequence was derived.
GENE DISCOVERY USING ZmDB
Within ZmDB, researchers have many avenues to gene discovery from different starting points. For example, our BLAST server can be used to compare a (non-maize) gene of interest with all the maize GSSs. From the displayed BLAST output page, researchers have the choice to perform further analyses using the integrated tools. For example, highly similar hits can be selected for multiple sequence alignment to find possible conserved domains between the gene of interest and other hits. Clicking the individual hit brings up the sequence display page. If pre-derived GSS to EST spliced alignments are displayed, this will indicate a likely active gene with the indicated (partial) exon–intron structure. If the hits include RescueMu-derived GSSs, then researchers can search the PhenotypeDB for mutant plants bearing the particular transposon insertion, which may provide direct clues to the function of the newly identified gene. A specialized GeneSeqer server allows spliced alignment with all plant ESTs which often allows accurate gene structure prediction even in the absence of cognate maize ESTs (12).
AVAILABILITY
ZmDB is accessible at the URL http://zmdb.iastate.edu. Data files and source code for some of the algorithms used at ZmDB can be downloaded from links on the home page. The manager of the database can be contacted by email at zmdb@iastate.edu.
Acknowledgments
ACKNOWLEDGEMENTS
ZmDB is supported by the USA National Science Foundation grant NSF#9872657, V.W. principal investigator. Team members include the authors plus Vicki Chandler, David Galbraith and Brian Larkins at the University of Arizona, Tucson; Sarah Hake at the University of California, Berkeley; Robert Schmidt and Laurie Smith at the University of California, San Diego; Marty Sachs at the University of Illinois, Urbana; and their collaborators at those locations.
REFERENCES
- 1.Gai X., Lal,S., Xing,L., Brendel,V. and Walbot,V. (2000) Gene discovery using the maize genome database ZmDB. Nucleic Acids Res., 28, 94–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walbot V. (1992) Strategies for mutagenesis and gene cloning using transposon tagging and T-DNA insertional mutagenesis. Annu. Rev. Plant Phys. Plant Mol. Biol., 43, 49–82. [Google Scholar]
- 3.Raizada M.N., Nan,G.-L. and Walbot,V. (2001) Somatic and germinal mobility of the RescueMu transposon in transgenic maize. Plant Cell, 13, 1587–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennetzen J.L., Chandler,V.L. and Schnable,P. (2002) National Science Foundation-Sponsored Workshop Report. Maize genome sequencing project. Plant Physiol., 127, 1572–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brendel V., Kurtz,S. and Walbot,V. (2002) Prospects and limits of comparative Arabidopsis—maize genomics. Genome Biol., 3, reviews 1005.1–1005.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandler V.L. and Brendel,V. (2002) Update: Maize genome sequencing project. Plant Physiol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang X. and Madan,A. (1999) CAP3: a DNA sequence assembly program. Genome Res., 9, 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Usuka J., Zhu,W. and Brendel,V. (1999) Optimal spliced alignment of homologous cDNA to a genomic DNA template. Bioinformatics, 16, 203–211. [DOI] [PubMed] [Google Scholar]
- 10.Kurtz S., Choudhuri,J.V., Ohlebusch,E., Schleiermacher,C., Stoye,J. and Giegerich,R. (2001) REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res., 29, 4633–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xing L. and Brendel,V. (2001) Multi-query sequence BLAST output examination with MuSeqBox. Bioinformatics, 17, 744–745. [DOI] [PubMed] [Google Scholar]
- 12.Brendel V. and Zhu,W. (2002) Computational modeling of gene structure in Arabidopsis thaliana. Plant Mol. Biol., 48, 49–58. [PubMed] [Google Scholar]