Abstract
Three-dimensional structures are now known within most protein families and it is likely, when searching a sequence database, that one will identify a homolog of known structure. The goal of Entrez's 3D-structure database is to make structure information and the functional annotation it can provide easily accessible to molecular biologists. To this end, Entrez's search engine provides several powerful features: (i) links between databases, for example between a protein's sequence and structure; (ii) pre-computed sequence and structure neighbors; and (iii) structure and sequence/structure alignment visualization. Here, we focus on a new feature of Entrez's Molecular Modeling Database (MMDB): Graphical summaries of the biological annotation available for each 3D structure, based on the results of automated comparative analysis. MMDB is available at: http://www.ncbi.nlm.nih.gov/Entrez/structure.html.
CONTENTS
Access
Molecular Modeling Database (MMDB) is Entrez's ‘Structure’ database (1). Querying by terms, for example, one may identify structures of interest based on a protein name. Links between databases provide other search mechanisms. A query of Entrez's MEDLINE® database, for example, can identify articles referring to a particular protein name. Links from this set of articles to ‘Structure’ may identify structures not found by direct query, since MEDLINE abstracts contains additional descriptive terms. At the time of writing, MMDB serves about 50 000 queries per day.
Data sources
Experimental 3D structure data are retrieved from the Protein Data Bank (2). Agreement of atomic coordinate and sequence data for each structure is checked and sequences are automatically modified, if necessary, to achieve exact agreement with coordinates. Data are mapped into a computer-friendly format encoded in ASN.1. This validation and encoding supports interoperable sequence, structure and alignment displays. MMDB currently contains about 20 000 structure entries, corresponding to about 40 000 chains and 70 000 3D domains.
Links, neighbours, and visualization
Sequences derived from MMDB are entered into Entrez's protein or nucleic acid sequence database, preserving a link to the corresponding structure. Links to MEDLINE are generated by citation matching (1). Links to Entrez's organism taxonomy database are validated manually (3). Sequence neighbours are identified by BLAST (4), and links to the Conserved Domain Database (CDD) by the reverse PSI-BLAST algorithm (5). Structure neighbours are identified by VAST (6). Entrez's integrated viewer, Cn3D (7), provides molecular-graphics visualization.
ANNOTATION
Structure summaries
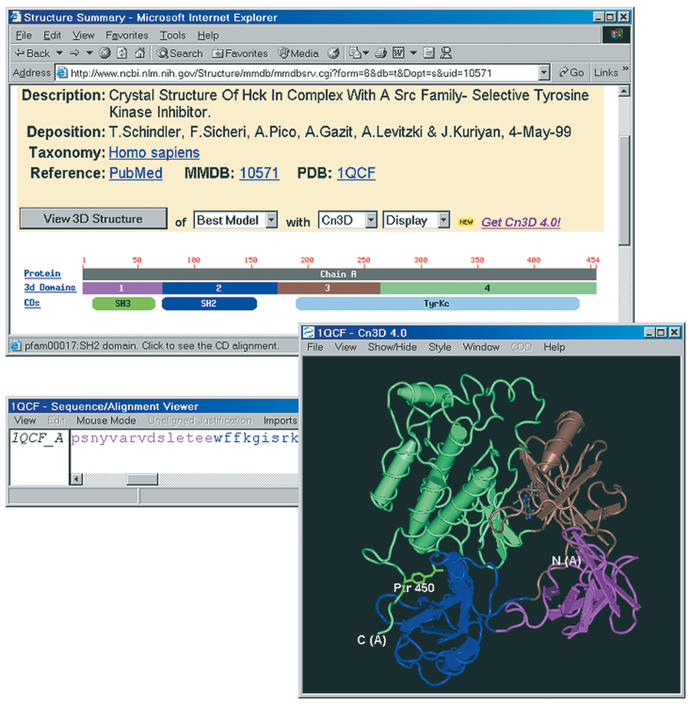
Entrez's ‘Structure summary’ provides a concise description of the contents of an MMDB entry and available annotation. Figure 1 presents an example, Hck Kinase, 1QCF (8). Links to MEDLINE and Taxon are provided together with descriptive text and a ‘View’ control to launch molecular-graphics visualization. The remainder of the display presents a graphical summary of macromolecular components. Each polypeptide (or polynucleotide) is described by a ‘sequence ruler’ that indicates chain lengths and the locations of protein domains. This graphical display links to annotation pertaining to individual chains and protein domains.
Figure 1.
MMDB's ‘structure summary’ for Hck kinase/1QCF. The location of the intra-molecular interaction of phosphotyrosine with the SH2 domain is highlighted in green.
MMDB employs two distinct but related definitions of protein domain. ‘3D domains’ are identified automatically as compact units within a polypeptide chain. As shown in Figure 1, colouring of 3D domains in the molecular graphics display matches that of the ‘boxes’ indicating their locations on the sequence ruler. 3D domains are the units for which automated structure neighbour calculations are performed, and the ‘box’ for each 3D domain (and complete chain) links to a display of its structure neighbours. A link to Entrez's text-listing of 3D domains is useful for advanced queries combining structural similarity with other attributes (3).
Entrez's CDD defines protein domains as recurrent evolutionary modules. In Figure 1, for example, a CDD ‘oval’ indicates that the region corresponding to the second 3D domain contains a member of the SH2 family. The SH2 ‘oval’ links to a detailed sequence/structure alignment, as predefined in CDD (5). Correspondence between 3D domains and conserved domains is not exact. The tyrosine kinase domain (‘TyrKc’) defined in CDD, for example, corresponds to two 3D domains, each representing a compact lobe in the structure.
STRUCTURE NEIGHBOURS
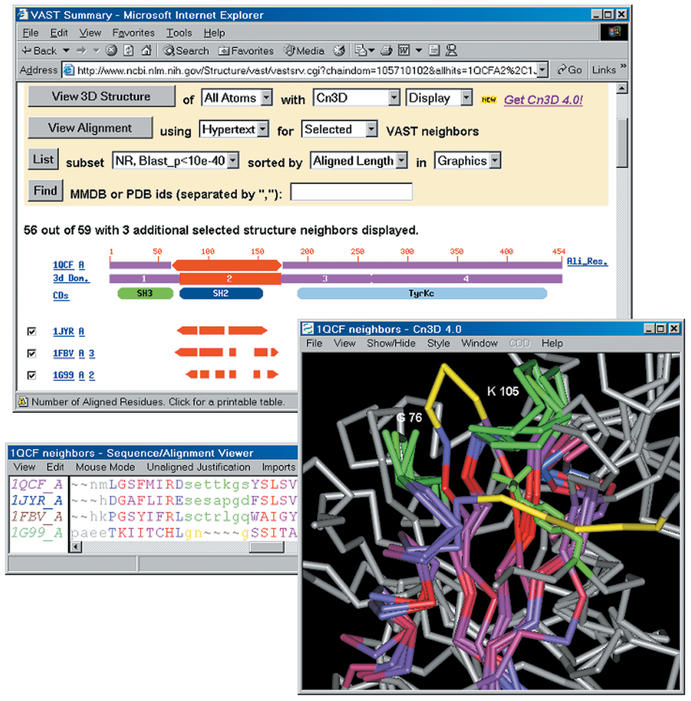
Structure neighbours are a rich source of biological annotation. Figure 2 shows an example, the structure neighbours of the SH2 domain of 1QCF. The structure of loop regions contributing to the intra-molecular phosphotyrosine binding site is preserved in 1JYR, a complex of Grb2 SH2 domain with a phosphotyrosine-containing peptide (9), and in 1FBV, a complex of c-Cbl with a phosphotyrosine-containing peptide (10). One may infer that proteins preserving this site are likely to bind phosphotyrosine. Consistent with this inference, structure 1G99, an Archaeal acetate kinase (11), does not preserve this site. If this protein shares a common ancestor with SH2 domains, it presumably belongs to a lineage that diverged prior to evolution of phophotyrosine binding. While superpositions based on 3D domains are normally adequate for structure-function analyses of this kind, Cn3D's alignment editing tools may be used to modify alignments and superpositions when necessary.
Figure 2.
MMDB's ‘VAST summary’ of selected structure neighbours of the SH2 domain (3D domain 2) of Hck kinase/1QCF. The locations of loop regions whose conformation is conserved in Grb2/1JYR and c-Cbl/1FBV are highlighted in green, as is the phosphotyrosine residue of a peptide bound to 1FBV. Analogous loop regions in acetate-kinase/1G99 are highlighted in yellow. The loop analogous to that near 1QCF K105 adopts a different conformation and the loop analogous to that near 1QCF G75 occludes the site where SH2 domains bind phosphotyrosine. Cn3D's alignment window displays the residues of 1QCF that may be superposed onto all selected neighbours; the degree of sequence conservation is indicated via a colour-ramp from blue-grey to red, with non-aligned sites shown in grey.
On average, there are over 600 structure neighbours for each 3D domain in MMDB. To help identify neighbours that provide useful annotation, Entrez's ‘VAST Summary’ provides a series of controls for selecting and sorting structure neighbours. As illustrated in Figure 2, the ‘alignment footprint’ of each neighbour indicates the region on the 3D domain serving as query that can be well superposed onto that neighbour. This display identifies structure neighbours similar to one another, where visualization of multiple-structure superpositions is informative. Other controls sort structure neighbours by measures of similarity and select subsets that include only one representative of sequence-similar subgroups. VAST-Search, which identifies neighbours of user-submitted structures, provides the same analysis tools.
FUTURE DIRECTIONS
Links to protein classifications like CDD are a valuable source of annotation, since descriptions and functional-site definitions are the result of expert curation. CDD alignments also identify the conserved core and in future we plan to use this information in sorting structure neighbours (12). Automated identification of sequence and structure neighbours provides the raw material for curated resources, however, and allows Entrez users to discover new relationships not yet described there. We plan to further improve tools for identification of informative sequence and structure neighbours.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the NIH Intramural Research Program for support. Questions should be addressed to: info@ncbi.nlm.nih.gov.
REFERENCES
- 1.Wheeler D.L., Church,D.M., Lash,A.E., Leipe,D.D., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Tatusova,T.A. and Wagner. (2003) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 31, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Westbrook J., Feng,Z., Jain,S., Bhat,T.N., Thanki,N., Ravichandran,V., Gilliland,G.L., Bluhm,W., Weissig,H., Greer,D.S., Bourne,P.E. and Berman,H.M. (2003) The Protein Data Bank: unifying the archive. Nucleic Acids Res., 31, 489–491.12520059 [Google Scholar]
- 3.Wang Y., Anderson,J.B., Chen,J., Geer,L.Y., He,S., Hurwitz,D.I., Liebert,C.A., Madej,T., Marchler,G.H., Marchler-Bauer,A., Panchenko,A.R., Shoemaker,B.A., Song,J.S., Thiessen,P.A., Yamashita,R.A. and Bryant,S.H. (2002) MMDB: Entrez's 3D-structure database. Nucleic Acids Res., 30, 249–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul S.F., Madden,T.L., Schaäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marchler-Bauer A., Anderson,J., Fedorova,N., DeWeese-Scott,C., Geer,L.Y., He,S., Hurwitz,D.I., Jackson,J.D., Jacobs,A., Lanczycki,C., Liebert,C., Liu,C., Madej,T., Marchler,G.A., Mazumder,R., Nikolskaya,A., Panchenko,A.R., Shoemaker,B.A., Song,J., Rao,R.B., Thiessen,P.A., Vasudevan,S., Wang,Y., Yamashita,R., Yin,J. and Bryant,S.H. (2003) CDD: A curated Entrez database of conserved domain alignments. Nucleic Acids Res., 31, 383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibrat J.F., Madej,T. and Bryant,S.H. (1996) Surprising similarities in structure comparison. Curr. Opin. Struct. Biol., 6, 377–385. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y., Geer,L.Y., Chappey,C., Kans,J.A. and Bryant,S.H. (2000) Cn3D: sequence and structure views for Entrez. Trends Biochem. Sci., 25, 300–302. [DOI] [PubMed] [Google Scholar]
- 8.Schindler T., Sicheri,F., Pico,A., Gazit,A., Levitzki,A. and Kuriyan,J. (1999) Crystal structure of Hck in complex with a Src family-selective tyrosine kinase inhibitor. Mol. Cell, 3, 639–648. [DOI] [PubMed] [Google Scholar]
- 9.Nioche P., Liu,W.Q., Broutin,I., Charbonnier,F., Latreille,M.T., Vidal,M., Roques,B., Garbay,C. and Ducruix,A. (2002) Crystal structures of the SH2 domain of Grb2: highlight on the binding of a new high-affinity inhibitor. J. Mol. Biol., 315, 1167–1177. [DOI] [PubMed] [Google Scholar]
- 10.Zheng N., Wang,P., Jeffrey,P.D. and Pavletich,N.P. (2000) Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell, 102, 533–539. [DOI] [PubMed] [Google Scholar]
- 11.Buss K.A., Cooper,D.R., Ingram-Smith,C., Ferry,J.G., Sanders,D.A. and Hasson,M.S. (2001) Urkinase: structure of acetate kinase, a member of the ASKHA superfamily of phosphotransferases. J. Bacteriol., 183, 680–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuo Y. and Bryant,S.H. (1999) Identification of homologous core structures. Proteins, 35, 70–79. [PubMed] [Google Scholar]