Abstract
The Conserved Domain Database (CDD) is now indexed as a separate database within the Entrez system and linked to other Entrez databases such as MEDLINE®. This allows users to search for domain types by name, for example, or to view the domain architecture of any protein in Entrez's sequence database. CDD can be accessed on the WorldWideWeb at http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=cdd. Users may also employ the CD-Search service to identify conserved domains in new sequences, at http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi. CD-Search results, and pre-computed links from Entrez's protein database, are calculated using the RPS-BLAST algorithm and Position Specific Score Matrices (PSSMs) derived from CDD alignments. CD-Searches are also run by default for protein–protein queries submitted to BLAST® at http://www.ncbi.nlm.nih.gov/BLAST.
CDD mirrors the publicly available domain alignment collections SMART and PFAM, and now also contains alignment models curated at NCBI. Structure information is used to identify the core substructure likely to be present in all family members, and to produce sequence alignments consistent with structure conservation. This alignment model allows NCBI curators to annotate ‘columns’ corresponding to functional sites conserved among family members.
INTRODUCTION
Protein domains are distinct units of protein three-dimensional structure, which also carry function. Proteins can be composed of single or multiple domains. As units of divergent molecular evolution, domains offer a rational level at which the protein universe may be studied. A few thousand conserved domain models are sufficient to cover more than two thirds of known protein sequences, greatly reducing the redundancy encountered in analyses involving sequence databases. Thus the annotation of protein sequence data with the location and extents of conserved domains has become an indispensable tool in the comparative analysis of genes and genomes. Pre-calculated assignments of functional and structural domains on protein sequence may provide valuable insights into the molecular evolution of single- and multiple-domain proteins, as well as help to validate other annotation.
We have continued to mirror two public collections of conserved domain models, Pfam (1) and SMART (2), and to convert their alignment models into searchable databases of Position Specific Score Matrices (PSSMs) (3). The current version of the CDD contains about 5000 such un-curated models, with sequence alignments imported from outside sources. In addition to this set, a first batch of several hundred curated alignment models is being offered.
NCBI-curated alignments are meant to give an accurate representation of the conserved core of a protein domain family, and can be used to instantiate approximate three-dimensional models for aligned sequences, assuming that the 3D structure of at least one family member is known. In some cases we need to resolve conflicts between imported sequence alignments and 3D structure information (4), such as the actual extents of conserved domains, the location and extents of conserved core blocks, and particular alignment details. Curated alignments are also meant to record conserved functional features, if applicable, in a way that assists visualization and permits computational transfer of such features across the family.
CDD CONTENTS
Access
CDD is an integral part of the Entrez data retrieval system (5), and can be accessed by querying or linking to Entrez's ‘Domains’ database. This allows retrieval by domain names and keywords found in functional descriptions. Conserved Domains are linked to the NCBI Taxonomy Database, PubMed®, and Entrez's protein database, which provides additional search mechanisms. A query of Entrez's PubMed Database, for example, may identify citations referring to a particular type of domain. Links from these citations to the ‘Domains’ database might help find the corresponding domain in CDD, as abstracts in PubMed often contain additional search terms not found in terse domain descriptions, and most entries in CDD are linked to relevant, carefully chosen citations.
We make extensive use of pre-calculated CD-Searches for proteins in Entrez. CDART, which stores that information, can be invoked from within Entrez to visualize domain architectures (6). Proteins in Entrez can now be neighboured by similar domain architecture, in addition to sequence similarity as detected by BLAST (7). Conserved Domains are neighboured to others by similarity, highlighting evolutionary relationships between families as well as the redundancy in the dataset, and by co-occurrence, highlighting domains, which are found next to each other in a set of protein sequences.
Data sources
Most of the domain models in CDD have been imported from two outside sources, Pfam and SMART. CDD also contains a small set of models labelled LOAD, and several hundred curated domain models, most of which are originally based on imported SMART and Pfam families. Some of the curated models have been generated de novo, to increase CDD coverage with respect to three-dimensional structures in MMDB (8). New SMART and Pfam distributions are imported on a regular basis, typically with several weeks delay. Upon import, we identify sequence fragments used in the alignments so that they can be linked to corresponding protein entries in Entrez. We also identify closely related three-dimensional structures in MMDB, so that alignment rows can be replaced with sequences corresponding to those structures. This allows us to present integrated sequence/structure/alignment views using Cn3D (9) as a helper application.
Links and Neighbours
Conserved Domains in Entrez are linked to PubMed citations, nodes in NCBI's taxonomy tree, and Entrez protein entries. PubMed identifiers are supplied by CDDs source databases, and those links are subject to change in curated CDs. For each domain alignment model, the set of representative sequences defines a common node in NCBIs taxonomy tree. Links to these common nodes are recorded in the database. The CDART database is the source for both links between CDs and proteins, and for CD neighbour data (6). CDART is populated with results from CD-searches comparing all of the proteins in Entrez to the current set of conserved domain models. CD-protein links are recorded as significant hits from these database searches, yielding E-values of 1e-2 or less. We record two types of CD–CD neighbour relationships. Two CDs are defined as similar if they hit overlapping intervals on a set of protein sequences. Two CDs are defined as co-occurring if they hit non-overlapping intervals on sets of protein sequences.
USING CDD TO FIND DOMAINS IN ENTREZ
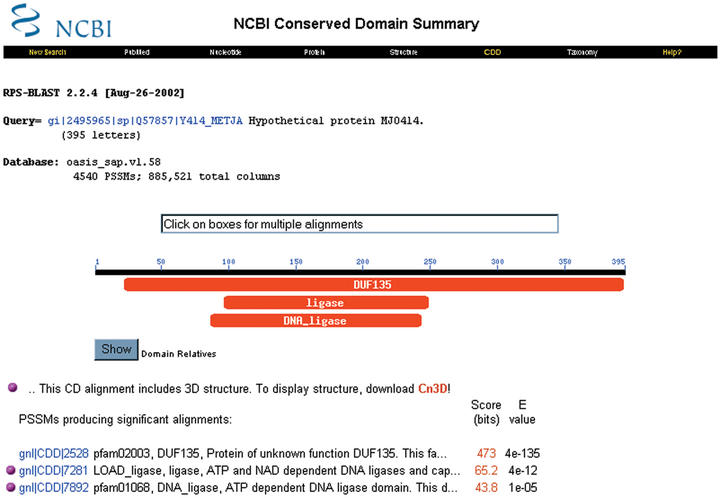
Users of NCBI's services are likely to encounter CDD in two ways. (i) When protein query sequences are submitted for BLAST searches against protein databases, the queries will be submitted to CD-Search by default, and the results—if any—will be displayed graphically on the intermediate BLAST results page. Clicking on the image will launch a browser window with the detailed results, which allow further analysis. (ii) Pre-calculated CD-search results exist for proteins in Entrez, and are readily available following the [Domains] link associated with protein records and document summaries. One might, for example, study a hypothetical protein from a complete genome sequence, say gi|2495965 from Methanocaldococcus jannaschii. Following the [Domains] link and expanding the summary to show more details will produce a graphical display, as shown in Figure 1. While the protein maps to a conserved family of unknown function (DUF135/pfam02003), the sequence also produces hits to two models for DNA ligases (pfam01068 and LOAD_ligase). In fact these three are grouped together with other domains as ‘related’ in the CDART database, as displayed on each member's conserved domain summary page. This bigger group of related domains comprises ATP- and NAD-dependent DNA Ligases, whose adenylation domains are known to share a well-conserved core structure around the active site (10). The representative model from the LOAD set, ‘LOAD_ligase’, aligns very diverse members from a large superfamily, also including RNA-ligases and mRNA capping enzymes (11).
Figure 1.
Pre-calculated CD-Search results are readily available for protein sequences in Entrez. Clicking on the coloured bars will launch alignment displays that merge the query into the domain alignment model, for further analysis. Domain annotation bars with identical colours have been grouped into sets of ‘related’ domains, indicating that they share many of the sequence intervals hit with significant E-values.
These and other interesting family relationships are recorded implicitly in CDD and CDART. Related domains share subsets of sequences for which overlapping intervals hit both domains with significant E-values in CD-Searches. The pre-recorded relationships help understand the redundancy in the imported and curated collections.
But how do we know whether these relationships are indicative of common molecular function? Multiple alignments are readily available for inspection, with the ability to colour by conservation. If a three-dimensional structure has been linked to the domain model, Entrez's structure viewer Cn3D can be used to interactively visualize structure and sequence data for a family. With these tools, and by exploring relevant literature, starting from CD-linked citations, the user may understand that it's in fact the catalytic core which is preserved among these families, and that they are likely to share a common enzymatic mechanism. However, if the location of functionally relevant residues had been recorded in the alignment models, it might have been easier to arrive at that conclusion.
MANUAL CURATION OF DOMAIN ALIGNMENT MODELS
Recording conserved features is one of the major tasks of expert domain alignment curation undertaken at NCBI. Alignment displays will highlight selected features, so that users can examine their agreement with residue conservation patterns, in particular when examining alignments with queries merged in according to CD-Search results. This may help, for example, to resolve the significance of CD-Search hits when the reported scores and E-values are not convincing.
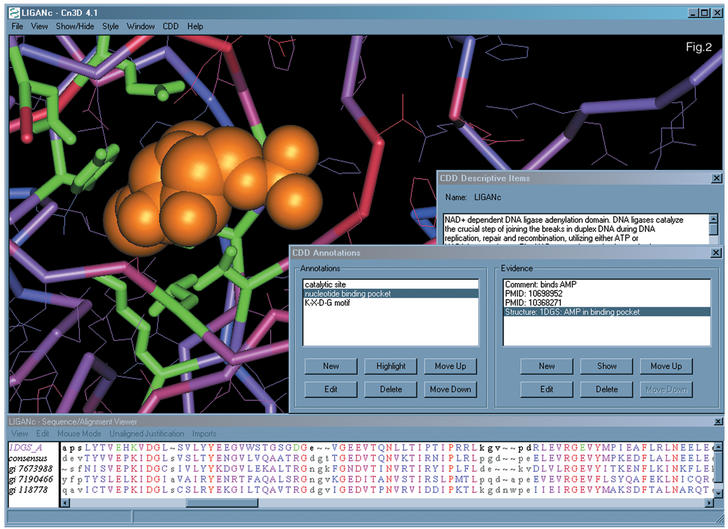
For selected features we record structure evidence, which can be visualized with Cn3D. Most commonly structure evidence is used with the annotation of sites involved in binding of cofactors, substrates, and other biopolymers, to indicate that we know about actual three-dimensional data sets demonstrating such molecular complexes. Figure 2 shows an example of such structure evidence.
Figure 2.
Visualization of ‘structure evidence’ for a feature recorded in CDD. The feature shown here is the nucleotide-binding pocket in cd00114 or LIGANc, the NAD-dependent subgroup of DNA ligases. The default colour scheme used by the WWW-server is green for residues, which are part of the feature evidence, and orange for heterogen groups.
Conserved features can be recorded only for residues aligned consistently across the family model. We find it necessary to re-evaluate and often change imported alignments to ensure this consistency. We also attempt to define the conserved core structure when curating alignments of diverse families, in agreement with data from comparative analysis of 3D structure (4).
We plan to update the contents of curated CDs periodically. The update process mines the CDART database for additional members of the domain family, which are dissimilar enough to already aligned rows to be interesting. Updates are curated, so that family membership as derived from the results of an automated procedure is validated. In the process of curating updates, new family members may suggest changes to the existing core model, for example, or provide additional data for feature annotation and feature evidence.
FUTURE DEVELOPMENTS
With data imported from a variety of sources, and with adding curated versions of many models to the collection, the set of conserved domains in CDD has become redundant. Relationships between conserved domains, as mentioned in the example above, may be interesting and lead to discovery, but they may as well just point out duplication of data. We intend to explicitly record relationships between curated domain models in CDD, by curating hierarchies of conserved domain models. Diverse families will be represented by ‘parent’ alignments with many divergent members, and very often it will be desirable to also represent more specific sub-families, for more precise functional annotation. If the resulting family relationships are recorded and clearly presented when visualizing results, users will be able to focus on the interesting aspects of data redundancy.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the NIH Intramural Research Program for support. We thank the authors of Pfam, SMART and LOAD, for creating invaluable resources and for helping with access to data. We are grateful towards the NCBI Blast group for developing RPS-BLAST and continuous support. Comments, suggestions, and questions are welcome and should be directed to: info@ncbi.nlm.nih.gov.
REFERENCES
- 1.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Letunic I., Goodstadt,L., Dickens,N.J., Doerks,T., Schultz,J., Mott,R., Ciccarelli,F., Copley,R.R., Ponting,C.P. and Bork,P. (2002) Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res., 30, 242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchler-Bauer A., Panchenko,A.R., Shoemaker,B.A., Thiessen,P.A., Geer,L.Y. and Bryant,S.H. (2002) CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res., 30, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marchler-Bauer A., Panchenko,A.R., Ariel,N. and Bryant,S.H. (2002) Comparison of sequence and structure alignments for protein domains. Proteins, 48, 439–446. [DOI] [PubMed] [Google Scholar]
- 5.Wheeler D.L., Church,D.M., Lash,A.E., Leipe,D.D., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Tatusova,T.A., Wagner,L. and Rapp,B.A. (2002) Database resources of the National Center for Biotechnology Information: 2002 update. Nucleic Acids Res., 30, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geer L.Y., Domrachev,M., Lipman,D.J. and Bryant,S.H. (2002) CDART: Protein Homology by Domain Architecture. Genome Res., 12, 1619–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J., Anderson,J.B., DeWeese-Scott,C., Fedorova,N.D., Geer,L.Y., He,S., Hurwitz,D.I., Jackson,J.D., Jacobs,A.R., Lanczycki,C.J., Liebert,C.A., Madej,T., Marchler-Bauer,A., Marchler,G.H., Mazumder,R., Nikolskaya,A.N., Rao,B.S., Panchenko,A.R., Shoemaker,B.A., Song,J.S., Thiessen,P.A., Vasudevan,S., Wang,Y., Yamashita,R.A., Yin,J.J. and Bryant,S.H. (2003) MMDB: Entrez's 3D-Structure Database. Nucleic Acids Res., 31, 474–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.http://www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml.
- 10.Singleton M.R., Hakannson,K., Timson,D.J. and Wigley,D.B. (1999) Structure of the adenylation domain of an NAD+-dependent DNA ligase. Struct. Fold. Des., 15, 35–42. [DOI] [PubMed] [Google Scholar]
- 11.Aravind L. and Koonin,E.V. (1999) Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol., 287, 1023–1040. [DOI] [PubMed] [Google Scholar]