Abstract
The DNA Data Bank of Japan (DDBJ, http://www.ddbj.nig.ac.jp) has collected and released more entries and bases than last year. This is mainly due to large-scale submissions from Japanese sequencing teams on mouse, rice, chimpanzee, nematoda and other organisms. The contributions of DDBJ over the past year are 17.3% (entries) and 10.3% (bases) of the combined outputs of the International Nucleotide Sequence Databases (INSD). Our complete genome sequence database, Genome Information Broker (GIB), has been improved by incorporating XML. It is now possible to perform a more sophisticated database search against the new GIB than the ordinary BLAST or FASTA search.
INTRODUCTION
The 50th anniversary of the discovery of the double helix (1) has immeasurable meanings in many disciplines including bioinformatics or information biology. This discovery was the first step in the invention of DNA sequencing (2,3) and then in the establishment of the International Nucleotide Sequence Databases (INSD), which is composed of DDBJ (http://www.ddbj.nig.ac.jp), EMBL Bank and GenBank. Boldly speaking, information biology was born and has advanced on two foundations: the establishment of INSD and the development of the computer tools for homology search (4–6). It can also be said that the scientific core of information biology is evolution (7), because DNA and proteins are products of organismic evolution over the past 4 billion years. Actually, this fact makes homology search possible and meaningful.
The collaboration among the three INSD members for more than 15 years has made it possible to provide an unlimited number of researchers worldwide with an ever-increasing amount of DNA sequence data and with the necessary computer tools for data retrieval and analysis. The amount of the data collected per year continues to steeply increase. Up till now, INSD all together has released more than 17 million entries or sequences to the public, and will perhaps do more than 30 million by the end of 2003. This means that the user has to spend more and more time on sequence retrieval or homology search against INSD. Therefore, it is desirable to improve the database search by use of more sophisticated retrieval methods than ordinary FASTA or BLAST search (5,6). One may also need to make a local database using the result of database search for further analysis on the smaller, more focused data set.
DDBJ is now at work on the incorporation of XML into its databases. We believe that use of XML will facilitate carrying out sophisticated retrievals, construction of a local database and integration with analysis tools in general. The user can now obtain the search results against DDBJ not only in the flat file format but also in the DDBJ-XML format. In this report, we will briefly describe our data collection/release in the past year and the application of XML to our whole genome database, GIB (8,9).
DATA COLLECTION/RELEASE AT DDBJ IN THE PAST YEAR
At the beginning of the last year (January 2001) large-scale human genome data from the Japanese human genome sequencing team was submitted to DDBJ. The entire human genome data set was then simultaneously published from the members of INSD on the internationally fixed date. After the release of the human genome data, the rate of data submission to DDBJ has not slowed down. Actually, the rate is even higher than that of the last year (2001). As of June 2002 (the DDBJ release 50) the total numbers of entries and bases collected and released by DDBJ are, respectively, 2,980,618 and 2,071,097,285. As to the international contribution of DDBJ at that time point, the proportions to the INSD totals are respectively 17.3% and 10.3%. DDBJ produced 68.7% more entries and 73.7% more bases in 2002 than in 2001. The corresponding figures for the year before the last (2000) are 42.6% and 51.6% (10). The contrast between the two pairs of figures indicates that the completion of human genome sequencing is expanding and accelerating whole genome sequencing in general, not otherwise.
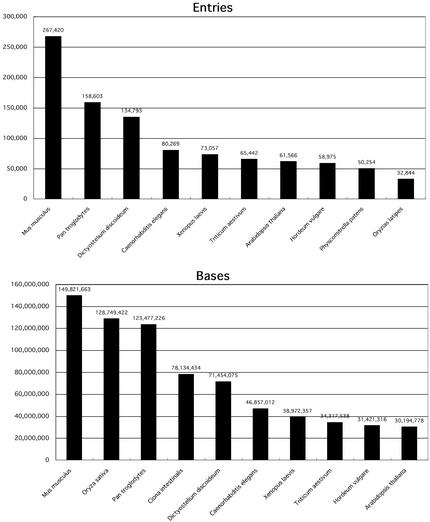
To see the increments over the past year in more detail, we picked the top ten species for which sequence data were submitted to DDBJ in that period. The increments for the top ten species are shown in Figure 1. At first glance, one notices that the species in the two sub-figures in Figure 1 do not coincide. In particular, rice (Oryza sativa), which is the second highest species in the number of bases, does not appear in the other figure, where the species are ranked in the number of entries. The reason for this is that the Japanese rice sequencing team has produced and submitted their data on a relatively small number of long sequences. The international rice genome sequencing consortium, which includes the Japanese team, plans to publish the whole rice genome sequence data by the end of 2002. The Japanese team is actively sequencing chromosomes 1, 2, 6, 7, 8 and 9 of that species.
Figure 1.
Top 10 species for which sequence data were submitted to DDBJ over the past year. Upper and lower figures respectively show the top 10 species in terms of the number of entries and that of bases. The vertical axis in the upper and lower figures respectively indicates the number of entries and that of bases. Number on the head of the bar indicates the exact number of entries or base for the species.
Mouse (Mus musculus) is the top species in both the numbers of entries and bases. This contribution was made by the Riken Hayashizaki team, which produced a tremendous amount of full length cDNAs (11). The mouse full length cDNA data are valuable for a number of research purposes. One such use is to elucidate the function of human genes by comparative genomics and experiment. It is generally accepted that the mouse genome has most of human homologues in it. In relation to the elucidation of the function of human genes, chimpanzee (Pan troglodytes) is certainly another target species, because it is the closest extant species to man. Pan troglodytes is ranked high in positions, both in the number of entries and in the number of bases. As an international chimpanzee genome sequencing consortium has been organized and activated by Japanese, Chinese, Korean, Taiwan and European teams, the data on the whole genome will be available in INSD in the near future. DDBJ is now in preparing for the collection of the chimpanzee genome data from the consortium. Another noteworthy species in Figure 1 is nemadoda (Caenorhabditis elegans). Sequence data for nemadoda have been submitted over the past several years by the Kohara team in the National Institute of Genetics.
APPLICATION OF XML TO THE DDBJ DATABASE
Before whole genome sequence data started to be submitted to INSD, one entry usually contained a sequence for one gene. In such a case, data presentation in the flat file format is convenient both to man and computer. However, the flat file format is not very efficient for genome sequence data, because one entry often contains more than one gene (perhaps a hundred or more), or a large number of feature items. This makes it difficult to extract the relevant pieces of information of one's interest from the entry. Therefore, we have itemized the pieces of information in an entry by XML, and devised the DDBJ-XML format for presenting the contents of an entry. The DDBJ-XML format is structurally simple, making the correspondence to the flat file straightforward.
XML can also be used as a script for describing ‘interface’ and ‘control’ of a given set of data or databases. We applied this property of XML to our Genome Information Broker to extend its function and facilitate its updating. GIB is a database for the complete genome sequences submitted to INSD. It mainly contains data for microbial genomes. At present, GIB provides the complete genome data for more than 90 species. Data for complete genomes are currently submitted to INSD at the rate of three to four species per month. Thus the number of species will soon exceed 100 and will continue to steadily increase. To cope with this situation, we have changed GIB from a single database to a set of parallel databases with the number of the databases being extendable by incorporating various mechanisms relating to XML. For example, we have made it possible to automatically transform a message in XML into SQL syntax so that search against GIB can be performed by a query in XML (9).
As a result, one can easily write a query in our XML format focusing on a particular item in an entry of GIB without paying attention to the relational table structure in the database. The XML format has changed many free form text fields into more structured XML tags, thus improving search efficiency. Therefore, one can avoid an unnecessary time-consuming text search. An example of a query and the retrieval result in XML is given in Figure 2. Using the element or tag in XML, one can combine keywords by logical ‘or’ and ‘and’ boolean operators in one's query. Another noteworthy point is that the retrieval result can be represented in various ways, making it easy to draw a graph, a pie chart or other figures. This is because each item of the result is represented in a tag pair. Also noted that the values are dynamically given during retrieval.
Figure 2.
Query and the retrieval result in XML. This example shows a query written for searching for virulence and membrane in Escherichia coli K12W3110 and the result both in XML. Note that the result is itemized in terms of species, chromosome, feature, qualifier and so forth.
We have developed an XML query system which is working towards makes it possible to integrate existing retrieval systems including relational database systems, homology and keyword search systems. This will make it possible for one to carry out a sophisticated database search against the new GIB by writing one's query in XML. For example, it is possible to conduct a homology search for a predefined region of a query sequence and obtain as a result not only the homologous sequences but also their feature information for the regions corresponding to the defined region in the query. One can further create a local database easily by incorporating the result of database search, because the result is given in a standard XML format. Such a complicated database search may not be possible by the ordinary BLAST or FASTA search.
CONCLUDING REMARKS
As a number of genome projects and consortiums have produced and submitted their data to INSD, INSD seems to operate now as ‘genome-wise’ rather than ‘gene-wise’ databases. This trend is natural in that the genes function interdependently in the networks of life from fertilization, to development, to birth, to growth, to aging and to death. Genome-wise understanding of a single gene will lead one to the elucidation of its comprehensive function. As a result, an entry of INSD tends to contain information on a large number of sequences, many pieces of features and qualifiers, and so forth. Our development of an XML query language will continue not only to alleviate the task of database search against such large and complex databases but also to facilitate database search for better results. It is also noted that the traditional gene-wise aspect of INSD should not be regarded as less important, because the function of a gene in the genome is often determined or deduced on the basis of that aspect.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the DDBJ members for making it possible to run DDBJ well in collaboration with EMBL Bank and GenBank. We are especially grateful to the Ministry of Education, Science, Culture, Sports and Technology for their enduring financial support. We also thank Ronald Taylor of the Center of Computational Biology, University of Colorado, for valuable comments.
REFERENCES
- 1.Watson J.D. and Crick,F.H.C. (1953) A structure for deoxyribose nucleic acids. Nature, 171, 737–738. [DOI] [PubMed] [Google Scholar]
- 2.Maxam A.M. and Gilbert,W. (1977) A new method for sequencing DNA. Proc. Natl Acad. Sci. USA, 74, 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanger F., Nicklen,S. and Coulson,A.R. (1977) DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Needlman S.B. and Wunsch,C.D. (1970) A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol., 48, 443–453. [DOI] [PubMed] [Google Scholar]
- 5.Lipman D.J. and Pearson,W.R. (1985) Rapid and sensitive protein similarity searches. Science, 227, 1435–1441. [DOI] [PubMed] [Google Scholar]
- 6.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 7.Tateno Y. and Gojobori,T. (1997) DNA Data Bank of Japan in the age of information biology. Nucleic Acids Res., 25, 14–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tateno Y., Fukami-Kobayashi,K., Miyazaki,S., Sugawara,H. and Gojobori,T. (1998) DNA Data Bank of Japan at work on genome sequence data. Nucleic Acids Res., 26, 16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fumoto M., Miyazaki,S. and Sugawara,H. (2002) Genome Information Broker (GIB): data retrieval and comparative analysis system for completed microbial genome and more. Nucleic Acids Res., 30, 66–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tateno Y., Imanishi,T., Miyazaki,S., Fukami-Kobayashi,K., Saitou,N., Sugawara,H. and Gojobori,T. (2002) DNA Data Bank of Japan (DDBJ) for genome scale research in life science. Nucleic Acids Res., 30, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium (2001) Functional annotation of a full-length mouse cDNA collection. Nature, 409, 685–690. [DOI] [PubMed] [Google Scholar]