Abstract
ArrayExpress is a new public database of microarray gene expression data at the EBI, which is a generic gene expression database designed to hold data from all microarray platforms. ArrayExpress uses the annotation standard Minimum Information About a Microarray Experiment (MIAME) and the associated XML data exchange format Microarray Gene Expression Markup Language (MAGE-ML) and it is designed to store well annotated data in a structured way. The ArrayExpress infrastructure consists of the database itself, data submissions in MAGE-ML format or via an online submission tool MIAMExpress, online database query interface, and the Expression Profiler online analysis tool. ArrayExpress accepts three types of submission, arrays, experiments and protocols, each of these is assigned an accession number. Help on data submission and annotation is provided by the curation team. The database can be queried on parameters such as author, laboratory, organism, experiment or array types. With an increasing number of organisations adopting MAGE-ML standard, the volume of submissions to ArrayExpress is increasing rapidly. The database can be accessed at http://www.ebi.ac.uk/arrayexpress.
BACKGROUND
Microarray experiments are generating a wealth of gene expression data, providing important insights into a variety of biological processes (1). To make maximum use of these data, a community infrastructure for sharing microarray data is needed (2). Three important elements of such an infrastructure are a data annotation standard, a common data exchange format, and public repositories for such data, analogous to DDBJ/EMBL/GenBank for molecular sequence data (3). The Minimum Information About a Microarray Experiment (MIAME)—a microarray data annotation standard (4) and Microarray Gene Expression Markup Language (MAGE-ML) (5)—an XML based data exchange format have been developed by the Microarray Gene Expression Data (MGED) society (http://www.mged.org) and Object Management Group (OMG, http://www.omg.org). ArrayExpress is a new public repository for microarray based gene expression data, which implements these standards.
ArrayExpress has three major goals: (i) to serve the scientific community as a repository for data that support publications, (ii) to provide the community with easy access to high quality gene expression data in a standard format, and (iii) to facilitate the sharing of microarray designs and experimental protocols. ArrayExpress accepts submissions in MAGE-ML format, or via a web-based submission tool, MIAMExpress. The ArrayExpress curation team assist data submitters in providing information required for MIAME compliance and curate the submitted data. The database can be accessed via a web-based query interface, and the data can be downloaded for local use or analysed via an online data analysis tool, Expression Profiler (6). Although ArrayExpress is still under development, all the components are functional, and new functionality is constantly being added. ArrayExpress has been accepting data submissions since February 2002. With an increasing number of microarray vendors and laboratories adopting the MAGE-ML and MIAME standards, the volume of submissions to ArrayExpress is growing rapidly.
DATA SUBMISSIONS
ArrayExpress accepts three types of submissions: arrays, experiments and protocols (including experimental and data processing protocols). Each of these can be submitted separately and is assigned a unique accession number. This can later be used as a reference, either within the database or externally. For example, researchers using a standard platform such as Affymetrix do not need to describe the array design, but should only reference the respective array accession number. An experiment submission must have relevant array designs and protocols linked. A journal publication may use ArrayExpress accession numbers to refer their supporting data.
There are two data submission routes to ArrayExpress: (i) directly via MAGE-ML files, or (ii) via a web-based submission interface, MIAMExpress, described below. As generation of MAGE-ML format data requires both a local Laboratory Information Management System (LIMS) and informatics support, this route is best suited for projects that have the necessary infrastructure (and is analogous to the route used by the sequencing centres for submissions to DDBJ/EMBL/GenBank). Currently, a MIAME compliant MAGE-ML based pipeline has been established with the Wellcome Trust Sanger Institute. Other similar pipelines, including ones from TIGR, Affymetrix, BASE, J-Express, Agileut, Rosetta Biosoftware and NCI, are under testing or construction. The MAGE-ML files should be submitted to a dedicated FTP site (for details see ArrayExpress home page).
MIAMExpress is a web-based tool, which allows users to annotate the submission either during, or upon the completion of the experiment. The current MIAMExpress Version 1.0 is a generic annotation tool, suitable for annotation of any microarray gene expression experiment, irrespective of organism or type. To use, MIAMExpress users need only an internet browser. The user creates an account and is presented with a series of web forms, which include a combination of drop-down fields (with appropriate controlled vocabularies) and free format text fields, to annotate the experiment. Tab-delimited data files are uploaded from the user's local computer and linked to the experiment submission. Arrays and protocols can also be submitted via MIAMExpress and can be linked to multiple experiments. Help is available from the curation team throughout the submission and contextual help is provided within the interface. Throughout the submission process, the data are stored in a submission database and are subsequently curated and then exported to ArrayExpress.
MIAMExpress is an open source project and consists of a perl-CGI interface, MySQL database, and MAGE-ML export component implemented using MAGEstk (5). The system can be installed locally and used as an ‘electronic notebook’ for microarray experiments, potentially allowing ‘one-click’ submissions to ArrayExpress or to any other database or tool that accepts MAGE-ML formatted data.
DATABASE QUERIES, DATA EXPORT AND ANALYSIS
Arrays, experiments and protocols can be queried by their accession numbers and also by a variety of parameters. For example, experiments can be queried on various parameters including author, laboratory, organism, experiment type (e.g. time series), experimental factors (e.g. compound) and details of the array used in the experiments (e.g. array manufacturer). On querying, a brief description (provided by the submitter) of all entries matching the query is returned. Complete information can be retrieved by selecting particular items from this list. A browsable list of the database content is also present in the query interface. Microarrays can contain tens or even hundreds of thousands of features. To explore array designs more easily, a generic tab-delimited display format has been designed, in which each row corresponds to a feature (spot) on the array.
The gene expression data can be exported in a tab-delimited format for downloading or for direct submission to Expression Profiler (6), a web-based tool for gene expression and other functional genomics data analysis. An expression analysis module, EPCLUST, allows further sub-selection and filtering of the data, management of multiple data sets, clustering and visualisation of these data, and performs similarity searches based on expression profiles. The URLMAP data cross-linking tool augments the EPCLUST analysis by linking to other tools and databases, e.g. metabolic pathway databases.
THE DATABASE DESIGN AND IMPLEMENTATION
ArrayExpress is an implementation of the Microarray Gene Expression Object Model (MAGE-OM), which has recently become an ‘available Specification’ of OMG standards group (http://cgi.omg.org/cgi-bin/doc?lifesci/01-10-01) and from which MAGE-ML was derived. The model and hence ArrayExpress are platform independent in terms of hardware and array platforms. The three major components: arrays, experiments and protocols are further described according to the MAGE-OM data model. ArrayExpress uses the MIAME definition of an experiment—a set of related hybridisations, data, protocols and array descriptions. There are several types of protocols, including extraction, labelling, hybridisation, scanning and data transformation protocols. Array design describes what occupies each feature (spot) on the array and provides the respective references to external databases.
For further information on the ArrayExpress design and implementation see (7). The latest database model, database scripts and application software are available from the ArrayExpress web site. ArrayExpress is implemented in Oracle; the query interface is implemented via Java servlets and uses Tomcat and Velocity.
FUTURE DEVELOPMENTS
ArrayExpress is an ongoing project and current developments focus on improving the query interface to exploit the full power of the MAGE-OM model. In particular, gene-centric queries combining data from several experiments will provide cross-platform analysis possibilities. This will be achieved via database warehousing, which also addresses the potential performance problems. ArrayExpress will be fully integrated with the relevant databases at the EBI and queries combining information from different databases will be possible.
The ontology developed by the MGED Ontology Working Group will be incorporated into future ArrayExpress query interfaces where possible. Future releases of MIAMExpress will incorporate terms from the MGED ontology and will also be used as a source of terms for the ontology. In addition, MIAMExpress will provide species or research area (e.g. toxicogenomics) specific interfaces, thus simplifying submissions for these data. Currently, we are developing toxicogenomic-specific and plant-specific interfaces as a part of collaborative projects. We intend to extend this to other areas, for example, those required by model organisms, where existing ontologies or controlled vocabularies will be used within the interface.
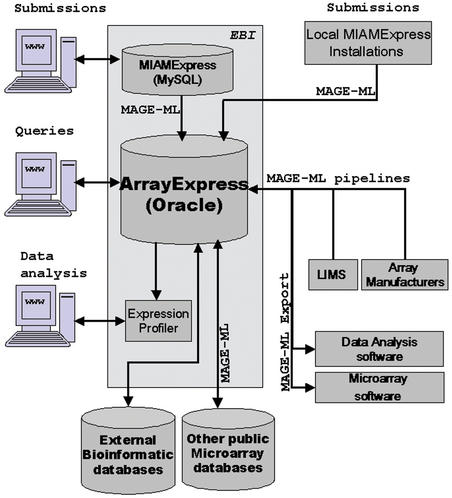
The infrastructure for data sharing is based on the adoption of the MAGE-ML data exchange format by the community, a process which is gathering momentum. In future as microarray LIMS support the use of MIAME and are able to export MAGE-ML, data submission to central repositories will become simpler (see Fig. 1). MAGE-ML is also an obvious candidate as a data exchange format between public repositories such as GEO at NCBI (8) or CIBEX currently under development at DDBJ. Moreover, the availability of common experimental and data processing protocols (described in a standard format) will encourage common laboratory practices. This, in turn, will serve to improve the comparability of datasets generated in different laboratories.
Figure 1.
A representation of the dataflow, database structure and MAGE-ML based infrastructure for data sharing.
In addition to the software related efforts described here, we are actively working with experimental centres and consortia to generate high quality MIAME compliant data. Examples of these include the toxicogenomics project coordinated by ILSI (http://www.ilsi.org) which is producing cross-platform gene expression data on the effects of various toxic compounds (9) and the cancer profiling project by the International Genomics Consortium (IGC) (10) who intend to screen thousands of tumour samples and deposit the data in ArrayExpress. The ArrayExpress team is interested in collaborating with all potential data providers and array manufacturers to establish direct MAGE-ML based pipelines for data and array design submissions to the database.
SUPPLEMENTARY INFORMATION
http://www.ebi.ac.uk/arrayexpress
ArrayExpress submission procedures.
Web-based database query interface.
ArrayExpress and MIAMExpress source code. http://ep.ebi.ac.uk/
Expression Profiler. http://www.mged.org/
MIAME.
Introduction to MAGE-OM and MAGE-ML data exchange format.
Ontology development for microarray experiments.
-
Recommendations on data processing.
http://cgi.omg.org/cgi-bin/doc?lifesci/01-10-01—the latest MAGE specifications
http://www.ii.uib.no/~bjarted/jexpress/—J-Express
http://jakarta.apache.org/tomcat/—Tomcat software
http://jakarta.apache.org/velocity/—Velocity software
http://www.perl.com/—Perl
http://www.mysql.com—MySQL
Acknowledgments
ACKNOWLEDGEMENTS
The ArrayExpress project is funded by EMBL, the European Commission (TEMBLOR grant), the EBI Industry Programme (Biostandards) and the International Life Sciences Institute (ILSI/HESI) toxicogenomics database grant. Initial funding was provided by Incyte and we particularly thank Lee Grower.
The authors would like to thank Rob Andrews, Jürg Bähler and Kate Rice (Sanger Institute), John Quackenbush and Joe White (TIGR), Paul Spellman (University of California at Berkeley) and Steve Chervitz (Affymetrix) all of whom who have generously provided their datasets and/or array designs in MAGE-ML format. We thank Tom Freeman (UK MRC-HGMP) for testing the MIAMExpress prototype. We acknowledge Jason Stewart (Open Informatics) for coordinating the development of the open source tools for processing MAGE-ML. We would also like to thank the Alan Robinson, MGED members and the entire EBI Microarray Informatics Team.
REFERENCES
- 1.The Chipping Forecast (1999) Suppl. Nature Genet., 21, 1–60. [Google Scholar]
- 2.Brazma A., Robinson,A., Cameron,G. and Ashburner,M. (2000) One-stop shop for microarray data. Nature, 403, 699–700. [DOI] [PubMed] [Google Scholar]
- 3.Stoesser G., Baker,W., van den Broek,A., Camon,E., Garcia-Pastor,M., Kanz,C., Kulikova,T., Leinonen,R., Lin,Q., Lombard,V., Lopez,R., Redaschi,N., Stoehr,P., Tuli,M.A., Tzouvara,K. and Vaughan,R. (2002) The EMBL Nucleotide Sequence Database, Nucleic Acids Res., 30, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brazma A., Hingamp,P., Quackenbush,J., Sherlock,G., Spellman,P., Stoeckert,C., Aach,J., Ansorge,W., Ball,C.A., Causton,H.C., Gaasterland,T., Glenisson,P., Holstege,F.C.P., Kim,I.F., Markowitz,V., Matese,J.C., Parkinson,H., Robinson,A., Sarkans,U., Schulze-Kremer,S., Stewart,J., Taylor,R., Vilo,J. and Vingron,M. (2001) Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nature Genet., 29, 365–371. [DOI] [PubMed] [Google Scholar]
- 5.Spellman P.T., Miller,M., Stewart,J., Troup,C., Sarkans,U., Chervitz,S., Bernhart,D., Sherlock,G., Ball,C., Lepage,M., Swiatek,M., Marks,W.L., Goncalves,J., Markel,S., Iordan,D., Shojatalab,M., Pizarro,A., White,J., Hubley,R., Deutsch,E., Senger,M., Aronow,B.J., Robinson,A., Bassett,D., Stoeckert,C.J.,Jr. and Brazma,A. (2002) Design and implementation of microarray gene expression markup language (MAGE-ML). Genome Biol., 3, research0046.1–0046.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vilo J., Kapushesky,M., Kemmeren,P., Sarkans,U. and Brazma,A. Expression Profiler. In Parmigiani,G., Garrett,E.S., Irizarry,R. and Zeger,S.L. (eds), The Analysis of Gene Expression Data: Methods and Software, Springer Verlag, New York, NY.
- 7.Brazma A., Sarkans,U., Robinson,A., Vilo,J., Vingron,M., Hoheisel,J. and Fellenberg,K. (2002) Microarray Data Representation, Annotation and Storage. Adv. Biochem. Eng. Biotechnol., 77, 113–139. [DOI] [PubMed] [Google Scholar]
- 8.Edgar R., Domrachev,M. and Lash,A. (2002) Gene Expression Omnibus: NCBI gene expression and hybridisation array data repository. Nucleic Acids Res., 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson D.E., Pettit,S.D. and Morgan,D.G. (2002) Use of genomics in mechanism based risk assessment. In Inoue,T. and Pennie,W.D. (eds), Toxicogenomics. Springer Verlag, Tokyo, pp. 194–203.
- 10.Knight J. (2001) Cancer comes under scrutiny in fresh genomics initiative. Nature, 4, 855. [DOI] [PubMed] [Google Scholar]