Abstract
WorfDB (Worm ORFeome DataBase; http://worfdb.dfci.harvard.edu) was created to integrate and disseminate the data from the cloning of complete set of ∼19 000 predicted protein-encoding Open Reading Frames (ORFs) of Caenorhabditis elegans (also referred to as the ‘worm ORFeome’). WorfDB serves as a central data repository enabling the scientific community to search for availability and quality of cloned ORFs. So far, ORF sequence tags (OSTs) obtained for all individual clones have allowed exon structure corrections for ∼3400 ORFs originally predicted by the C. elegans sequencing consortium. In addition, we now have OSTs for ∼4300 predicted genes for which no ESTs were available. The database contains this OST information along with data pertinent to the cloning process. WorfDB could serve as a model database for other metazoan ORFeome cloning projects.
INTRODUCTION
A nearly complete version of the Caenorhabditis elegans genome sequence was released in December 1998 (1). With its ∼19 000 predicted protein-encoding open reading frames (ORFs), C. elegans is one of the first multicellular model organisms for which functional genomic and proteomic approaches became feasible (2,3). Among these approaches, those relying upon the availability of a DNA fragment for each gene analyzed have been the fastest to develop. For example, global expression profiling using microarray technologies (4) and comprehensive functional analysis using RNA interference (RNAi) can be performed with a partial genomic fragment for each predicted ORF (5–8). Other approaches are more directly focused on the proteins encoded by predicted ORFs. For example, a protein–protein interaction mapping project has been initiated for C. elegans (9–11) and several groups have embarked on the resolution of large numbers of protein structures (12). In addition, one can expect the development in the near future of C. elegans protein chips (13), biochemical genomics (14) and other proteomic approaches (15,16). Such protein-based high-throughput approaches are all highly dependent upon the availability of a complete set of cloned ORFs or ‘ORFeome’. Initiated in 1998, the primary goal of the C. elegans ORFeome cloning project is to generate this resource using a cloning system which allows subsequent transfer of the ORFs into multiple expression vectors (9,17). Another important goal is to help with annotating the large number of predicted ORFs for which insufficient expressed sequence tag (EST) information is available to confirm GeneFinder predictions (17).
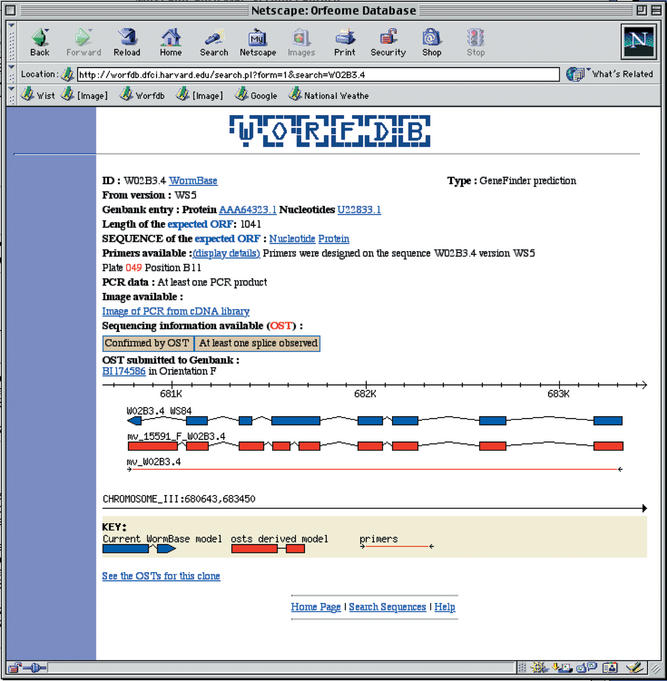
WorfDB (Worm ORFeome DataBase) was created to integrate the data from the ORFeome cloning project (17). In the first version of the cloned ORFeome (worm ORFeome v1.0) recently completed (J. Reboul in preparation) ∼12 000 ORFs have been successfully cloned. Each clone is available as a pool of Escherichia coli transformants and as a sequence-verified DNA preparation. WorfDB enables the scientific community to search for availability and quality of the clones. Every clone has been sequenced from both the 5′ and 3′ end, generating ∼28 000 ORF Sequence Tags (OSTs). The analysis of these OSTs has allowed the correction of ∼3400 predicted ORFs and gives experimental evidence for the expression of ∼4300 genes for which no ESTs were available. This sequencing information should be valuable to the C. elegans community as well as the scientific community at large because it provides a better annotation of the C. elegans proteome. The database currently contains sequence data for (i) all predicted ORFs from WormBase versions WS5 to WS9 (2); (ii) all GenBank entries submitted by the C. elegans research community; (iii) cDNAs submitted by the transcriptome analysis project (J. Thierry-Mieg in preparation). All of the data pertinent to the cloning process is stored in the database. This information includes the name and sequence of the primers used to amplify the ORFs from our worm cDNA library, the coordinates of the primers in their 96 well storage plates, photographs of agarose gels displaying the resulting PCR products, and any sequencing information available on the clones. A graphic display allows the visualization of the differences between the latest ORF structure prediction from WormBase and the OST structure obtained by the ORFeome project (Fig. 1).
Figure 1.
An example of a sequence page. The user can follow links to other database and visualize the photograph of an agarose gel displaying the corresponding PCR product. The bottom of the page shows the difference between the current WormBase gene model and the model derived from OST sequencing.
DESCRIPTION OF THE DATABASE
The database is a relational database using the SQL database server MySQL (T.c.X) with a web interface developed using perl and DBI. The sequence data have been extracted primarily from WormBase and GenBank using in-house extraction tools. The primers were designed automatically using OSP (18) and entered in the database with a link to their sequences and a reference to their position in their corresponding 96 well plate. Pictures of the corresponding PCR products analyzed by electrophoresis and ethidium bromide staining were entered one at a time with a reference to the primer plates. The OSTs obtained from sequencing the clones were aligned using the program ACEMBLY (ftp://ftp.ncbi.nlm.nih.gov/repository/acedb/ACEMBLY) and the alignment information extracted as gff files. The graphic display is implemented using the perl modules Bio::Graphics and Bio::DB::GFF and uses data from a gff database containing the gff dump files from WormBase merged with the gff files extracted from ACEMBLY.
SEARCHING THE DATABASE
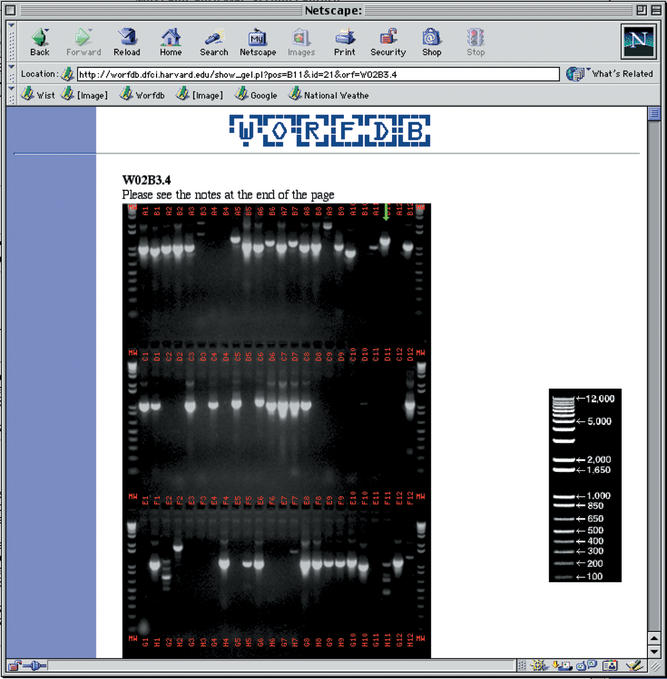
The database can be queried in two ways, either through a simple form with which one can search for clone, gene or protein names, or through a blast interface that queries all the sequences in the ORFeome database. The database contains alias and deprecated names that are linked to current names. In cases in which a search identifies more than one database entry, the names of all corresponding hits are displayed for the user to choose. If hits correspond to sequences that have the same name but have been modified between versions of WormBase or GenBank, only the most recent version is displayed with a note to the user that an older version exists. The sequence is presented in one page that gives links to other relevant databases (WormBase, GenBank) as well as a link to the ORFeome data available for this sequence (Fig. 1). If available, the image of the PCR products can be visualized by the user. The image is processed to highlight the position of the ORF of interest on the gel (Fig. 2). These images are valuable to assess the presence of multiple bands that can suggest the presence of alternative splice forms. WorfDB can also be queried through an LDAS (Lightweight Distributed Annotation System) server (http://worfdb.dfci.harvard.edu/cgi-bin/das/orfeome) that contains the annotation distributed in WormBase merged with our OSTs that are defined as ‘partial_cds’.
Figure 2.
An example of a photograph of one of the ∼220 agarose gels present in the database. The PCR product for the ORF of interest is highlighted with a green arrow pointing to the correct lane.
FUTURE OF WorfDB
We have now started the identification and archiving of full-length wild type clones for each ORF. This process is allowing the identification of large numbers of differentially spliced variants. As this information is accumulated in the process of analysis, it will be added to the database. In addition, our lab aims to define a protein interaction map for all the C. elegans proteins. The ORFeome database should serve as a good foundation to manage the data that we are generating. Ultimately, this database will be integrated with our Interaction map database (9). The data from the ORFeome cloning are shared with the WormBase database and reciprocal links are present on each of the databases and this interconnectivity should be increased in the future.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Laurent Jacotot for his help and Troy Moore, Jim Hartley, Mike Brasch, Hongmei Lee, Lynn Doucette-Stamm for reagents and sequencing. This work was supported by grants 5R01HG01715-02 (NHGRI and NIGMS), 7 R33 CA81658-02 (NCI) and 232 (MGRI) awarded to M.V.
REFERENCES
- 1.The C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 2.Sternberg P.W. (2001) Working in the post-genomic C. elegans world. Cell, 105, 173–176. [DOI] [PubMed] [Google Scholar]
- 3.Vidal M. (2001) A biological atlas of functional maps. Cell, 104, 333–339. [DOI] [PubMed] [Google Scholar]
- 4.Reinke V., Smith,H.E., Nance,J., Wang,J., Van Doren,C., Begley,R., Jones,S.J., Davis,E.B., Scherer,S., Ward,S. and Kim,S.K. (2000) A global profile of germline gene expression in C. elegans. Mol. Cell, 6, 605–616. [DOI] [PubMed] [Google Scholar]
- 5.Fraser A.G., Kamath,R.S., Zipperlen,P., Martinez-Campos,M., Sohrmann,M. and Ahringer,J. (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature, 408, 325–330. [DOI] [PubMed] [Google Scholar]
- 6.Gonczy P., Echeverri,G., Oegema,K., Coulson,A., Jones,S.J., Copley,R.R., Duperon,J., Oegema,J., Brehm,M., Cassin,E., Hannak,E., Kirkham,M., Pichler,S., Flohrs,K., Goessen,A., Leidel,S., Alleaume,A.M., Martin,C., Ozlu,N., Bork,P. and Hyman,A.A. (2000) Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature, 408, 331–336. [DOI] [PubMed] [Google Scholar]
- 7.Maeda I., Kohara,Y., Yamamoto,M. and Sugimoto,A. (2001) Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol., 11, 171–176. [DOI] [PubMed] [Google Scholar]
- 8.Piano F., Schetter,A.J., Mangone,M., Stein,L. and Kemphues,K.J. (2000) RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr. Biol., 10, 1619–1622. [DOI] [PubMed] [Google Scholar]
- 9.Walhout A.J.M., Sordella,R., Lu,X., Hartley,J.L., Temple,G.F., Brasch,M.A., Thierry-Mieg,N. and Vidal,M. (2000) Protein interaction mapping in C. elegans using proteins involved in vulval development. Science, 287, 116–122. [DOI] [PubMed] [Google Scholar]
- 10.Davy A., Bello,P., Thierry-Mieg,N., Vaglio,P., Hitti,J., Doucette-Stamm,L., Thierry-Mieg,D., Reboul,J., Boulton,S., Walhout,A.J., Coux,O. and Vidal,M. (2001) A protein–protein interaction map of the Caenorhabditis elegans 26S proteasome. EMBO Rep., 2, 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boulton S.J., Gartner,A., Reboul,J., Vaglio,P., Dyson,N., Hill,D.E. and Vidal,M. (2002) Combined functional genomic maps of the C. elegans DNA damage response. Science, 295, 127–131. [DOI] [PubMed] [Google Scholar]
- 12.Chance M.R., Bresnick,A.R., Burley,S.K., Jiang,J.S., Lima,C.D., Sali,A., Almo,S.C., Bonanno,J.B., Buglino,J.A., Boulton,S., Chen,H., Eswar,N., He,G., Huang,R., Ilyin,V., McMahan,L., Pieper,U., Ray,S., Vidal,M. and Wang,L.K. (2002) Structural genomics: a pipeline for providing structures for the biologist. Protein Sci., 11, 723–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu H., Klemic,J.F., Chang,S., Bertone,P., Casamayor,A., Klemic,K.G., Smith,D., Gerstein,M., Reed,M.A. and Snyder,M. (2000) Analysis of yeast protein kinases using protein chips. Nature Genet., 26, 283–289. [DOI] [PubMed] [Google Scholar]
- 14.Martzen M.R., McCraith,S.M., Spinelli,S.L., Torres,F.M., Fields,S., Grayhack,E.J. and Phizicky,E.M. (1999) A biochemical genomics approach for identifying genes by the activity of their products. Science, 286, 1153–1155. [DOI] [PubMed] [Google Scholar]
- 15.Ho Y., Gruhler,A., Heilbut,A., Bader,G.D., Moore,L., Adams,S.L., Millar,A., Taylor,P., Bennett,K., Boutilier,K., Yang,L., Wolting,C., Donaldson,I., Schandorff,S., Shewnarane,J., Vo,M., Taggart,J., Goudreault,M., Muskat,B., Alfarano,C., Dewar,D., Lin,Z., Michalickova,K., Willems,A.R., Sassi,H., Nielsen,P.A., Rasmussen,K.J., Andersen,J.R., Johansen,L.E., Hansen,L.H., Jespersen,H., Podtelejnikov,A., Nielsen,E., Crawford,J., Poulsen,V., Sorensen,B.D., Matthiesen,J., Hendrickson,R.C., Gleeson,F., Pawson,T., Moran,M.F., Durocher,D., Mann,M., Hogue,C.W., Figeys,D. and Tyers,M. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature, 415, 180–183. [DOI] [PubMed] [Google Scholar]
- 16.Gavin A.C., Bosche,M., Krause,R., Grandi,P., Marzioch,M., Bauer,A., Schultz,J., Rick,J.M., Michon,A.M., Cruciat,C.M., Remor,M., Hofert,C., Schelder,M., Brajenovic,M., Ruffner,H., Merino,A., Klein,K., Hudak,M., Dickson,D., Rudi,T., Gnau,V., Bauch,A., Bastuck,S., Huhse,B., Leutwein,C., Heurtier,M.A., Copley,R.R., Edelmann,A., Querfurth,E., Rybin,V., Drewes,G., Raida,M., Bouwmeester,T., Bork,P., Seraphin,B., Kuster,B., Neubauer,G. and Superti-Furga,G. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 17.Reboul J., Vaglio,P., Tzellas,N., Thierry-Mieg,N., Moore,T., Jackson,C., Shin-i,T., Kohara,Y., Thierry-Mieg,D., Thierry-Mieg,J., Lee,H., Hitti,J., Doucette-Stamm,L., Hartley,J.L., Temple,G.F., Brasch,M.A., Vandenhaute,J., Lamesch,P.E., Hill,D.E. and Vidal,M. (2001) Open-reading-frame sequence tags (OSTs) support the existence of at least 17 300 genes in C. elegans. Nature Genet., 27, 332–336. [DOI] [PubMed] [Google Scholar]
- 18.Hillier L. and Green,P. (1991) OSP: a computer program for choosing PCR and DNA sequencing primers. PCR Methods Appl., 1, 124–128. [DOI] [PubMed] [Google Scholar]