Abstract
We have developed a bacterial “two-hybrid” system that readily allows selection from libraries larger than 108 in size. Our bacterial system may be used to study either protein–DNA or protein—protein interactions, and it offers a number of potentially significant advantages over existing yeast-based one-hybrid and two-hybrid methods. We tested our system by selecting zinc finger variants (from a large randomized library) that bind tightly and specifically to desired DNA target sites. Our method allows sequence-specific zinc fingers to be isolated in a single selection step, and thus it should be more rapid than phage display strategies that typically require multiple enrichment/amplification cycles. Given the large library sizes our bacterial-based selection system can handle, this method should provide a powerful tool for identifying and optimizing protein–DNA and protein–protein interactions.
Selection and screening methods are powerful tools for studying macromolecular interactions. Examples of such methods include the yeast-based one-hybrid and two-hybrid systems (for studying protein–DNA and protein–protein interactions, respectively) and bacterial-based phage display methods (for studying either type of interaction). These systems have been used to identify interaction partners for particular DNA or protein targets, and they also have been used in combination with mutagenesis or randomization strategies to study the details of biologically important interactions (for reviews, see refs. 1–5). The development of bacterial-based systems analogous to the yeast one-hybrid and two-hybrid methods could, in principle, facilitate the rapid analysis of larger libraries (due to the higher transformation efficiency and faster growth rate observed with Escherichia coli). Such methods also might be faster than phage display, which is an enrichment technique requiring multiple rounds of affinity purification and amplification (e.g., see ref. 6) and may allow studies of sequences (or much larger proteins) that are not readily displayed on a phage surface.
Several bacterial one- and two-hybrid systems have been proposed, but there have been no reports in which these actually have been used to identify candidates from a real library (reviewed in ref. 7). This may reflect practical limitations with these existing systems. Most of these methods actually are designed as genetic screens (8–10) and thus cannot be readily used with libraries larger than ≈105–106 in size. Two genetic selection systems have been proposed for studying protein–protein interactions, but neither method is readily adaptable to the analysis of protein–DNA interactions (11, 12).
In this report, we describe the design and testing of an E. coli-based selection method that can detect either protein–DNA or protein–protein interactions and that can handle libraries larger than 108 in size. We tested our method by selecting Cys2His2 zinc finger variants similar to those previously isolated by phage display (6, 13). The results of our selection, the rapidity of our method, and the versatility of the underlying transcriptional activation scheme suggest that this bacterial-based system should provide a useful tool for identifying and characterizing protein–DNA and protein–protein interactions.
Materials and Methods
Selective Medium.
HIS-selective medium is composed of M9 minimal medium supplemented with 10 μM ZnCl2, 10 μg/ml thiamine, 200 μM adenine, 50 μg/ml carbenicillin, 30 μg/ml chloramphenicol, 30 μg/ml kanamycin, 50 μM isopropyl β-d-thiogalactoside, 20 mM 3-aminotriazole (3-AT), and 17 aa (all except histidine, methionine, and cysteine). For HIS-selective medium plates, agar was added to a final concentration of 1.5%, and the concentration of carbenicillin was increased to 100 μg/ml.
Plasmids and Bacterial Strains.
The αGal4 protein used in this study contains residues 1–248 of the E. coli RNA polymerase α subunit fused (by an Ala-Ala-Ala linker) to residues 58–97 of the yeast Gal4 protein. The pACYC184-derived plasmid pACL-αGal4 expresses αGal4 from a tandem, IPTG-inducible lpp/lacUV5 promoter.
The Gal11P-Zif123 fusion protein contains residues 263–352 of the yeast Gal11P protein (with a N342V mutation; ref. 14) fused by a 9-aa linker Ala-Ala-Ala-Pro-Arg-Val-Arg-Thr-Gly to residues 327–421 of Zif268 (the region encoding the three zinc fingers). The phagemid pBR-GP-Z123 expresses the Gal11P-Zif123 hybrid protein from an IPTG-inducible lacUV5 promoter. The pBR-GP-Z12BbsI phagemid is analogous to pBR-GP-Z123 except that Zif finger 3 is replaced with a modified Zif finger 1 in which the sequence encoding residues −1 through 6 of the finger recognition helix is replaced by unrelated sequence (a “stuffer” fragment) flanked by BbsI restriction sites. All phagemids used in this study can be easily “rescued” from cells by infection with a filamentous helper phage; infectious phage particles produced by these cells contain single-stranded phagemid DNA.
The reporter construct that expresses HIS3 (Pzif-HIS3-aadA) has the Zif268-binding site sequence 5′-GCGTGGGCG-3′ centered at base pair −63 relative to the transcription start site of a weak E. coli lac-promoter derivative (the Pwk promoter). The three selection strain reporters change the zinc finger binding site of Pzif-HIS3-aadA, replacing the sequence 5′-TCGACAAGCGTGGGCG3-′ (bases −74 to −59 relative to the transcription start site) with sequences that should allow binding of the desired zinc finger variants: 5′-CAAGGGTTCAGGGGCG3-′ (for nuclear receptor element, NRE), 5′-GGCTATAAAAGGGGCG3-′ (for TATA), or 5′-TGGGACATGTTGGGCG3-′ (for p53). Each of these reporters was transferred (by recombination) to an F′-episome encoding lacIq repressor and then introduced into strain KJ1C in a single step essentially as described (ref. 15; J.K.J. and C.O.P., unpublished data). The resulting strains then were each transformed with the pACL-αGal4 plasmid to create the NRE, TATA, p53, and Zif selection strains.
E. coli strain KJ1C, which has a deletion in the hisB gene, was constructed as follows: Strain SB3930 (F- ΔhisB463) was transduced to tetracycline resistance with P1vir phage grown on strain JCB40 (F- Δ(gpt-proAB-arg-lac)XIII zaj∷Tn10). Tetracycline-resistant colonies were screened for pro-, arg- lac-, and his- phenotypes.
Randomized Zinc Finger Library.
The zinc finger variant library was constructed by cassette mutagenesis. Randomized oligonucleotides synthesized by using a two-column method (16) were ligated to BbsI-digested pBR-GP-Z12BbsI vector (replacing the stuffer fragment in this phagemid) to create a library of zinc finger variants. Each member of this library has three zinc fingers: two constant fingers (fingers 1 and 2 of Zif268) and a third, carboxy-terminal finger (also derived from finger 1 of Zif268) in which recognition helix residues −1, 1, 2, 3, 5, and 6 are randomized. Our randomization scheme allowed 24 possible codons, encoding 19 possible amino acids (no cysteine) and one stop codon. The sequence complexity of the resulting library is ≈2 × 108. This ligation was electroporated into E. coli XL-1 Blue cells (Stratagene) and yielded >109 transformants. These were pooled, amplified, and then infected with VCS-M13 helper phage (Stratagene) to yield a high titer stock of phage harboring single-stranded versions of the phagemid library.
Selection Protocols.
For initial selections with each of the three variant sites, >1010 selection strain cells were infected with ≈109 ampicillin-resistance transducing units of phage from the phagemid library. After recovery under nonselective conditions for 1.5 h, infected cells were plated at a density of ≈1 to 5 × 108 ampicillin-resistant colonies/plate on HIS-selective medium. (Control experiments indicated a false positive rate of ≈3 × 10−8 under these selection conditions.) The largest surviving colonies were retested for growth on HIS-selective medium plates supplemented with 60 μg/ml spectinomycin (we chose 80–90 colonies for the NRE and TATA selections and 240 colonies for the p53 selection). Candidates that regrew on these plates then were chosen for phagemid-linkage testing.
The second NRE selection was performed in two stages, in an attempt to isolate additional variants. In the first stage, >1010 NRE selection strain cells were infected with ≈6 × 109 ampicillin-resistance transducing units of phage from the phagemid library. After recovery under nonselective conditions, the infection was plated at a density of ≈6 × 108 ampicillin-resistant colonies/plate on HIS-selective medium. One-half of the ≈900 surviving colonies were pooled and amplified in liquid HIS-selective medium supplemented with 50 μg/ml spectinomycin. This pooled culture was infected with VCS-M13 helper phage, grown overnight in 2× YT medium (1.6% tryptone, 1% yeast extract, 0.5% NaCl) supplemented with 50 μg/ml spectinomycin, and a high titer stock of phage was isolated. For the second stage, fresh NRE selection cells were infected with phage containing the enriched library of phagemids (from the first stage), and these were plated on HIS-selective medium plates. Twenty-four surviving colonies of various sizes were retested for growth on HIS-selective medium plates (supplemented with 60 μg/ml spectinomycin) and these then were checked for phagemid linkage.
Phagemid-Linkage Testing.
Colonies that grew on HIS-selective medium then were tested to see whether survival was phagemid-linked. Candidates were inoculated into liquid HIS-selective medium supplemented with 100 μg/ml spectinomycin (but lacking 3-AT). All of the NRE and TATA selection candidates, and the 72 fastest-growing p53 selection candidates, were infected with VCS-M13 helper phage, and the resulting phage-containing supernatants were harvested. Each candidate phage was used to infect fresh selection strain cells (corresponding to those on which it was originally selected), and these infected cells were plated on HIS-selective medium. Growth under these conditions demonstrates that activation of HIS3 expression is linked to the presence of the phagemid (and thus suggests that the phagemid-encoded zinc fingers bind to the DNA target subsite on which they were selected).
Binding Site Preference Testing.
To test the ability of the selected zinc fingers to discriminate among different binding sites, recovered phagemids were introduced (by phage infection) into NRE, p53, TATA, and Zif selection strain cells. Infected cells were plated on HIS-selective medium and growth was scored qualitatively after 24 h growth at 37°C and 18 h continued growth at room temperature. Under these conditions, we have found that survival of a selection strain indicates that the variant finger can bind the target subsite present on the reporter. If a zinc finger variant permits selection strains (other than the one in which it was initially isolated) to survive on selective medium, this suggests that the variant finger binds semispecifically or nonspecifically.
Sequencing of Candidates.
To prepare candidates for sequencing, the phage stocks of clones with a phagemid-linked phenotype were used to infect XL-1 Blue cells. Plasmid DNA was isolated from these cells (Qiagen, Valencia, CA) and used for dideoxy sequencing.
Results
An Improved E. coli-Based Two-Hybrid Selection System for Studying Protein–DNA and Protein–Protein Interactions.
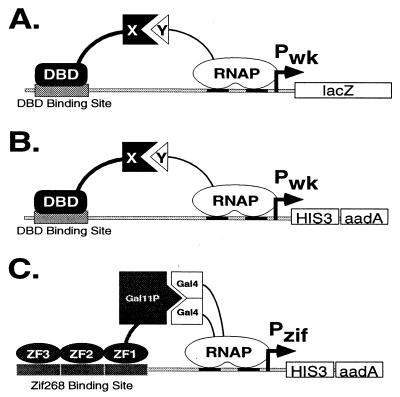
To design a bacterial-based selection method for studying protein–DNA and protein–protein interactions, we began with an existing genetic screen previously developed by Hochschild and coworkers (7, 8, 10). In this screen, as in the yeast two-hybrid system, there are two fusion proteins that interact in a way that leads to transcriptional activation of a lacZ-reporter gene (Fig. 1A). One protein is composed of a DNA-binding domain (DBD) fused to another domain represented as X in Fig. 1A. The second protein contains the domain Y fused to a subunit of the E. coli RNA polymerase. In this arrangement, activation of lacZ expression requires appropriate protein–DNA and protein–protein interactions: The DBD must bind to a DNA-binding site (DBS) positioned near the promoter, and domain X must simultaneously interact with domain Y to recruit RNA polymerase to the promoter, thereby activating transcription. The major advantage of this system is that almost any protein–DNA (DBD–DBS) or protein–protein (X–Y) interaction should mediate transcriptional activation. However, because lacZ is used as a reporter gene in this system, candidates must be identified by a visual phenotype (e.g., their blue color on 5-bromo-4-chloro-3-indolyl β-d-galactoside plates). Thus, the system (in this form) cannot readily be used to screen libraries larger than ≈105–106 in size.
Figure 1.
(A) Transcriptional activation in a previously described E. coli-based genetic screen [developed by Hochschild and coworkers (8, 10)] for studying protein–DNA and protein–protein interactions. (B) Modified reporter template for our E. coli-based genetic selection system. (C) Model for transcriptional activation of the Pzif promoter by fusion proteins Gal11P-Zif123 and αGal4. ZF1, ZF2, and ZF3 are the three zinc fingers of the Zif268 protein. (Although Gal11P-Zif123 efficiently activates our Pzif promoter, we note that the spacing between the Zif268-binding site and the transcription start site has not yet been optimized.)
To improve this previously described system so that it can be used to analyze libraries larger than 108 in size, we replaced the lacZ gene used in the Hochschild genetic screen with the selectable yeast HIS3 gene (Fig. 1B). HIS3 encodes an enzyme required for histidine biosynthesis that can complement the growth defect of E. coli cells bearing a deletion in the homologous hisB gene (ΔhisB cells) (17, 18). In addition, 3-AT, which is a competitive inhibitor of HIS3, can be used to titrate the level of HIS3 expression required for growth on medium lacking histidine (19). (Thus, in the presence of 3-AT, a higher level of activation is required to allow growth on selective medium.) We find that HIS3 is attractive for use with large libraries because: (i) >108 ΔhisB cells harboring a HIS3 gene expressed from the Pwk promoter can be plated on a regular-size Petri dish containing HIS-selective medium, and (ii) we find that these cells have a very low false positive rate (≈3 × 10−8) on HIS-selective medium (data not shown).
Our modified construct also contains the bacterial aadA gene (which confers resistance to the antibiotic spectinomycin) (20) positioned just downstream of the HIS3 gene (Fig. 1B). We refer to this construct as the Pwk-HIS3-aadA operon because Pwk directs coordinated expression of the HIS3 and aadA genes (data not shown). Although selection for increased aadA expression is not suitable for direct analysis of large libraries (we find this allows a relatively high background breakthrough; data not shown), we used spectinomycin in certain steps to maintain selective pressure (see Materials and Methods).
Zinc Finger Domains Can Bind DNA and Activate Transcription in E. coli.
We tested our E. coli-based system by applying it to a problem previously studied using phage display: the selection, from a large randomized library, of zinc finger variants with altered DNA-binding specificities (for review, see ref. 21). However, before proceeding with selections, we first examined whether a wild-type zinc finger protein could bind DNA and activate transcription in our system. (Relatively little information was available on the activity of Cys2His2 zinc finger proteins in bacteria.) To do this, we constructed fusion proteins containing fragments of the yeast Gal11P and Gal4 proteins that previously had been shown to interact with each other (10, 14). Thus, we fused a Gal11P fragment to the three zinc fingers of the murine Zif268 protein (creating the Gal11P-Zif123 protein), and we replaced the carboxyl-terminal domain of the E. coli RNA polymerase α subunit with a Gal4 fragment (creating the chimeric αGal4 protein). A Zif268 DNA-binding site was positioned upstream of our Pwk-HIS3-aadA operon to create the Pzif-HIS3-aadA operon (Fig. 1C), and this cassette was introduced into a ΔhisB E. coli strain in single copy to create the Zif-reporter strain.
We then tested whether the Gal11P-Zif123 and αGal4 proteins could work together as a two-hybrid system to activate transcription of the Pzif-HIS3-aadA operon. We found that Zif reporter strain cells expressing only the αGal4 protein do not grow on HIS-selective medium, but the same cells can grow when the Gal11P-Zif123 protein is expressed together with the αGal4 protein. We also found that activation requires all three Zif268 fingers: a Gal11P fusion protein that contains only the first two zinc fingers from Zif268 does not permit growth on selective medium. These results indicate that the Gal11P-Zif123 and αGal4 proteins can work together to activate transcription in our E. coli system. We presumed that the DNA-bound Gal11P-Zif123 acts by recruiting (or stabilizing) RNA polymerase complexes that have incorporated αGal4. These results also give some information about the DNA-affinity threshold for activation because we found that fingers 1 and 2 of Zif268 alone are not sufficient.
Selection Strategy for Isolation of Zinc Finger Variants.
Because our initial results indicated that zinc fingers could function in E. coli and that our activation scheme worked as expected, we proceeded to test our system by isolating zinc finger variants from a large randomized library. We chose target DNA subsites that had been used in an earlier phage display study (6, 13). This previous study had involved selecting zinc finger variants that would bind to sequences normally recognized by important eukaryotic DNA-binding proteins. The AAA target subsite used in our experiments is part of a TATA box, the TGT target subsite is part of a p53-binding site, and the TCA target subsite is part of an NRE. We refer to these sequences as the TATA, p53, and NRE target subsites.
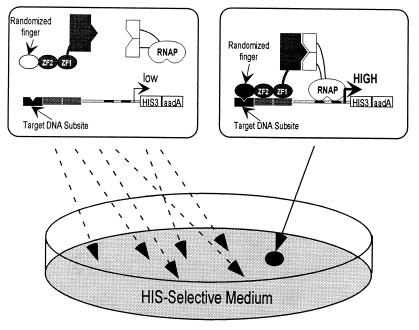
Our strategy for identifying variant zinc fingers that bind specifically to a particular “target” DNA subsite relies on the ability of our system to distinguish between zinc finger proteins that bind using two fingers (recognizing 6–7 bp) from those that bind using three fingers (recognizing 9–10 bp). We synthesized a large library of three-finger Zif268 derivatives (each fused to the Gal11P fragment). In this library, the first two fingers of Zif268 remain constant, but the recognition helix of the third, carboxyl-terminal finger is randomized (see Materials and Methods). We also prepared selection strains with the appropriate zinc finger binding sites upstream of the Pwk-HIS3-aadA operon. [Each of these has the normal binding subsites for fingers 1 and 2 of Zif268, but the third subsite (black notched rectangle, Fig. 2) is changed to include one of the target DNA subsites of interest (AAA for TATA; TGT for p53; TCA for NRE).] Each of these ΔhisB selection strains also contain a plasmid expressing the αGal4 protein, and these bacteria are referred to as the TATA, p53, and NRE selection strains. [As a control for use in binding site specificity studies (see below), we also constructed a corresponding Zif selection strain that has an intact Zif268-binding site (containing subsites for all three Zif268 fingers) positioned upstream of the Pwk promoter.]
Figure 2.
An E. coli-based selection system for identifying zinc finger variants from large randomized libraries. (Left) A selection strain cell bearing a randomized zinc finger (white oval) that is unable to bind the target DNA subsite of interest (black box). This candidate fails to activate transcription of the weak promoter controlling HIS3 expression and therefore cells expressing this candidate fail to grow on HIS-selective medium. (Right) A library candidate bearing a particular zinc finger (one member of the randomized library) (black oval) that can bind the target DNA site. This candidate can activate HIS3 expression and therefore cells expressing this candidate grow on HIS-selective medium.
To perform a selection with one of these three target subsites, we introduced >5 × 108 members of the phagemid library into the appropriate selection strain and plated the cells on HIS-selective medium. From our earlier controls, we expected that growth would require three functional fingers; thus, a cell should survive only if it happens to express a protein with a finger that binds tightly to the target subsite (Fig. 2).
Positive candidates identified on HIS-selective medium then were checked in several ways: Each candidate was first tested to verify that the phenotype of growth on selective medium was linked to the phagemid encoding the zinc finger library candidate (phagemid-linkage test, see Materials and Methods). Clones that still appeared positive then were tested to see how well they distinguish among the NRE, TATA, p53, and Zif subsites (binding site preference test, see Materials and Methods). Finally, clones were sequenced to determine which amino acids were preferred at the positions that had been randomized.
Selection of Zinc Fingers That Bind the TATA Target Subsite.
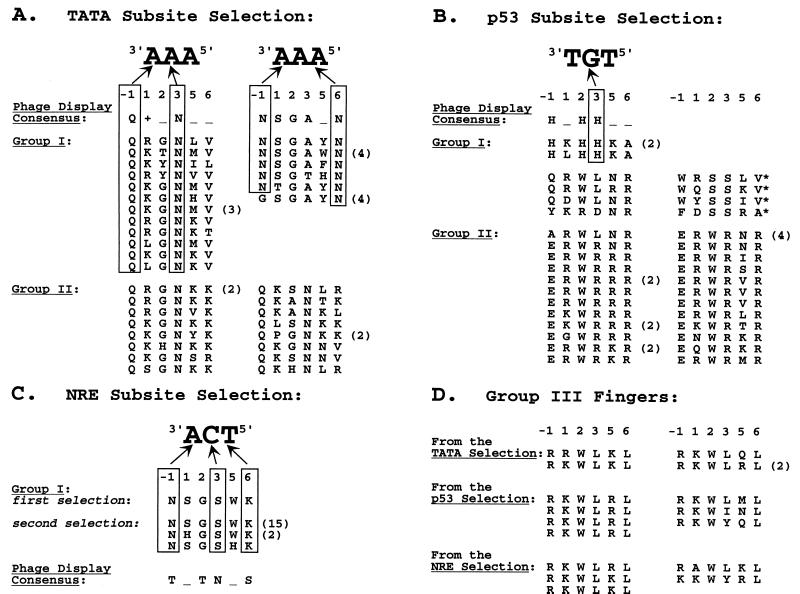
From the ≈5 × 108 zinc finger variants introduced into the TATA selection strain, we identified 49 candidates with a phagemid-linked phenotype. Based on their ability to distinguish among the TATA, p53, NRE and Zif subsites, these candidates could be categorized into three groups. Group I candidates bound specifically to the TATA target subsite. Group II candidates bound semispecifically (with a strong preference for the TATA subsite over the Zif subsite); group III candidates bound nonspecifically to all four subsites tested (with a preference for the Zif and p53 subsites over the TATA and NRE subsites). Amino acid sequences are shown in Fig. 3A (groups I and II) and Fig. 3D (group III) and reveal striking conserved patterns for each of the groups.
Figure 3.
Recognition helix sequences of fingers isolated by our selection. For candidates that were isolated multiple times (as judged by nucleotide sequence), the number of clones obtained is shown in parentheses. The consensus sequence(s) of fingers selected by phage display for each target subsite also are shown (6). +, positively charged residue; _, no discernible preference; *, candidates with a 2-bp deletion downstream of the sequence encoding the recognition helix; and arrows illustrate a few of the most plausible potential base contacts.
Selection of Zinc Fingers That Bind the p53 Target Subsite.
From the ≈1.3 × 109 zinc finger variants introduced into the p53 selection strain, we identified 48 candidates that demonstrate a phagemid-linked phenotype. Based on their ability to distinguish among the four different subsites, these candidates could be categorized into three groups. Group I candidates bound specifically to the p53 target subsite. Group II candidates bound semispecifically (with a general preference for the p53 subsite over the Zif subsite); group III candidates bound nonspecifically to all four subsites tested (again with a slight preference for the Zif and p53 subsites over the TATA and NRE subsites). The amino acid sequences of the recognition helices of these candidates are shown in Fig. 3B (groups I and II) and Fig. 3D (group III). Striking patterns of conserved residues were seen in each group.
Selection of Zinc Fingers That Bind the NRE Target Site.
Approximately 2 × 109 zinc finger variants were introduced into the NRE selection strain, and we obtained two candidates that demonstrated a phagemid-linked phenotype. One candidate binds specifically to the NRE target subsite (and also exhibits very weak binding to the TATA subsite). The second candidate binds nonspecifically to all four subsites tested (with a preference for the Zif and p53 subsites over the NRE and TATA subsites).
To isolate additional clones that recognize the NRE subsite, we performed a modified two-stage selection procedure. In the first stage, we repeated the selection for the NRE subsite and pooled 50% of the surviving colonies (≈450 candidates). In the second stage, finger-encoding phagemids isolated from this enriched pool (see Materials and Methods) then were reintroduced into the NRE selection strain and plated again on selective medium. All 24 colonies chosen for further analysis displayed a phagemid-linked phenotype, and these zinc fingers could be categorized into two groups on the basis of their observed specificities. Group I sequences bound well to the target NRE subsite (with very weak binding to the TATA subsite). Group III candidates bound nonspecifically to all four subsites tested (with a preference for the Zif and p53 subsites over the NRE and TATA subsites). The recognition helix sequences of all of the selected candidates are shown in Fig. 3C (group I) and Fig. 3D (group III). As with our other selections, striking patterns of conserved residues were observed in each of these groups.
Discussion
Selection of Variant Zinc Fingers with Altered DNA-Binding Specificities by Using a Bacterial-Based Selection Method.
Our bacterial-based selection system was designed to rapidly identify and characterize protein–DNA and protein–protein interactions. To test our method, we performed selections to identify variant zinc fingers that would bind selectively to desired target DNA subsites. We discuss these results in some detail in the following paragraphs, but our main observation was that the affinity and specificity of the selected fingers seems comparable, if not superior, to those obtained in earlier phage display studies (which required multiple rounds of selection and amplification).
For the TATA selection, subsite-specific fingers identified by our method (TATA group I) defined two consensus sequences, and these closely matched the two consensus sequences observed in fingers isolated by phage display (Fig. 3A). However, the randomization scheme used in constructing our library allowed aromatic amino acids (Phe, Tyr, and Trp) that were not represented in the codon scheme used for the corresponding phage display library (6, 13). One consensus sequence obtained with our selection appeared to specify an aromatic residue at position 5 of the recognition helix (NSGAθN, where θ is an aromatic residue). The corresponding phage display-derived consensus (NSGA_N) did not define any particular class of residues at this position. Our selection also yielded another class of fingers that appear to be semispecific for the TATA subsite (TATA group II fingers). The sequences of these fingers also matched one of the phage display consensus sequences, but all (except one) of these semispecific fingers were distinguishable from the specific fingers (TATA group I) by the presence of either an asparagine at position 5 or a positively charged residue at position 6 (Fig. 3A). Thus, the results for this subsite are quite clear: our selection yielded fingers that bind specifically to the TATA subsite, and the sequences of these fingers match well with those isolated by phage display.
For the p53 selection, we isolated a number of fingers that bind specifically to the intended target subsite (p53 group I). The recognition helix sequences of two of these fingers match the consensus sequence of those obtained by phage display (Fig. 3B). We note that the remaining p53 group I fingers have an aromatic residue at either position −1 or 2 of the recognition helix and thus would not have been present in libraries used for earlier phage display experiments. In addition, fingers isolated by our method that bind semispecifically to the p53 subsite (p53 group II fingers) all possess a tryptophan at position 2. Although the nature of some of the sequence-specific contacts made by these fingers is unclear, the conservation of specific aromatic residues at certain positions suggests an important role in DNA recognition. Again, our results with this subsite are very encouraging: our selection yielded a number of fingers that bind specifically to the p53 target subsite. Some of these fingers match the consensus obtained by phage display whereas others suggest that aromatic residues may play an important role in zinc finger-DNA recognition.
For the NRE target subsite, an initial attempt using our selection method yielded only one finger (NSGSWK) that bound preferentially to the target sequence. Based on our existing knowledge of zinc finger-DNA recognition (reviewed in ref. 21), one can postulate reasonable contacts between recognition helix residues of this finger and bases in the primary strand of the NRE subsite (Fig. 3C). However, we initially were concerned by the relatively low frequency of fingers selected for this site, and we repeated the selection using an additional enrichment step in an attempt to isolate more fingers. The great majority of sequences isolated this way had the same amino acid sequence as the candidate originally selected (NSGSWK) but two closely related sequences (NSGSHK and NHGSWK) also were identified. These results suggested that we might have obtained a small number of clones merely because very few candidates in our library can pass the threshold set in our NRE selection.
As shown in Fig. 3C, the sequences of fingers isolated in our NRE selections do not match the consensus sequence for fingers selected by phage display. We performed several experiments to explore the basis of this difference: We first checked our library by sequencing random candidates to ensure that there was no drastic bias in nucleotide distribution and were able to rule this out as a plausible explanation (unpublished data). We then decided to directly introduce (in exactly the same context) one of the fingers (TRTNKS) that had been selected by phage display (6) and see whether it could work in our system as a Gal11P-zinc finger fusion protein. We found that NRE selection strain cells expressing this TRTNKS finger fusion protein grew very poorly on HIS-selective medium whereas the same cells expressing the NSGSWK finger fusion (obtained in our selections) grew robustly (unpublished data). The simplest explanation for this result is that the TRTNKS finger fusion bound poorly to the NRE subsite and therefore only weakly stimulated HIS3 expression. This explanation is supported by our observation that earlier selections with the NRE subsite, using a prototype of our system in which zinc fingers were expressed from a much higher copy number phagemid, had yielded the TRTNKS as well as the NSGSWK finger (J.K.J. and C.O.P., unpublished data). This suggests that our current system sets a very stringent standard for the NRE selections and may account for why we isolated such a small number of specific candidates.
We also used our binding site preference assay to compare the specificity of the NSGSWK finger we had selected for the NRE subsite with that of the TRTNKS finger selected by phage display. In our bacterial-based assays, the NSGSWK finger bound specifically to the NRE subsite and bound only very weakly to the TATA subsite. By contrast, the TRTNKS finger bound only weakly to all four subsites (exhibiting a preference for the NRE and TATA subsites over the p53 and Zif subsites) (unpublished data). These results suggest that the NSGSWK finger we selected actually binds more tightly and specifically in our system than the TRTNKS finger identified earlier by phage display.
Each of our three selections also yielded a small percentage of fingers that bind nonspecifically to all four DNA subsites tested. Surprisingly, all of these fingers match a consensus sequence of the form R+WL+L (where + denotes a positively charged residue, Fig. 3D). These fingers are rich in positive charge and may make extra phosphate contacts. We also note that all of these fingers have a tryptophan residue at position 2 and thus would not have been present in the libraries used for earlier phage display experiments. This highly conserved set of nonspecific fingers raises many interesting questions: What level of specificity is required for a zinc finger protein to function in our assay (and thus to what extent does the E. coli chromosome function as a nonspecific competitor)? How do these fingers bind? Why is this particular class of nonspecific fingers the only type selected in our system?
In summary, the TATA and p53 subsite selections demonstrate that our bacterial-based system can isolate fingers similar to those obtained previously by phage display. Only a few fingers were obtained in the NRE subsite selections, but it appears that these may actually bind with better affinity and specificity than those obtained by phage display. Most significantly, we believe our method offers a more rapid alternative to phage display because it permits functional fingers to be isolated in a single selection step instead of using multiple rounds of enrichment. We also note that (as with recent phage display efforts from this lab and other laboratories) we took no special precautions to perform our selections in an anaerobic environment. We envision that our rapid bacterial-based system will be particularly useful for projects requiring multiple zinc finger selections (performed either in parallel or sequentially).
General Strategies for Studying Protein–DNA and Protein–Protein Interactions by Using Our Bacterial-Based Two-Hybrid Selection System.
This report demonstrates that our bacterial-based system can be used in a manner analogous to the yeast one-hybrid method to identify variant zinc fingers that bind to a specific DNA subsite. We also have found that a number of other eukaryotic DBDs can readily function in our system (J. Miller, J. Kanter, J.K.J., E.I.R., and C.O.P., unpublished results). Thus, we expect that our method also could be readily used to identify DNA-binding proteins from cDNA libraries or random peptide libraries. (Note: When planning selections for DNA-binding proteins, one may need to take account of methylation activity. The known activites of DNA adenine methyltransferase, DNA cytosine methyltransferase, and EcoKI methyltransferase should not be a problem for any of our current sites. However, in other cases it may be desirable to use E. coli strains defective for a particular methylase.)
With a few minor modifications, our selection method also could be used to identify and study protein–protein or protein–peptide interactions. In this application (analogous to the yeast two-hybrid method), the protein target (the “bait” or domain Y in Fig. 1 A and B) could be fused to either the dimeric α subunit or to the monomeric ω subunit of RNA polymerase. The protein or peptide library to be analyzed (the “prey” or domain X in Fig. 1 A and B) could be fused to either a dimeric (e.g., bacteriophage λcI protein) or monomeric (e.g., Zif268) DBD. [Previous experiments have shown that different interacting proteins X and Y can effect transcriptional activation and that the magnitude of this activation correlates well with the strength of the X-Y interaction (reviewed in ref. 22)]. The reporter in this application would be the Pwk-HIS3-aadA operon bearing an upstream binding site for the particular DBD used in the experiment. As with other applications of our system, the phagemid rescue feature simplifies and reduces the time required to test plasmid linkage and to analyze interaction specificity.
Our bacterial-based selection system offers a number of potentially significant advantages over analogous yeast-based one- and two-hybrid methods (reviewed in ref. 7). In particular it offers the ability to analyze libraries larger than 108 in size, faster growth rate, greater potential permeability to small molecules (23), the absence of a requirement for nuclear localization, and the possibility of studying proteins that are toxic when expressed in yeast. Unlike phage display methods, our selection system permits the single-step isolation of candidates in an in vivo context and also bypasses complications that may be associated with export of proteins to the cell membrane (as required to display on the phage surface). This report demonstrates that a bacteria-based two-hybrid system can be used successfully to identify candidates of interest from a large library (>108 in size). Our HIS3-based system provides a rapid selection method with a low false positive rate, and it can easily be titrated to be more or less stringent simply by varying the concentration of 3-AT inhibitor in the medium. Our method also is amenable to high-throughput analysis and automation, as many steps are performed in a 96-well format. We envision that our genetic selection method will provide a powerful, broadly applicable tool for identifying and characterizing both protein–DNA and protein–protein interactions.
Acknowledgments
We thank Scot Wolfe for the gift of the randomized library oligos, the E. coli Genetic Stock Center for strain SB3930, Ann Hochschild and Simon Dove for providing precursor strains and plasmids, Alan Frankel, Scot Wolfe, and Jeff Miller for comments on the manuscript, Scot Wolfe, Jeff Miller, Bryan Wang, and Kevin Struhl for discussions, and Tim Jenkins for help with preliminary experiments. J.K.J. is supported by a Howard Hughes Medical Institute Physician Postdoctoral Fellowship. C.O.P. is supported by the Howard Hughes Medical Institute.
Abbreviations
- DBD
DNA-binding domain
- NRE
nuclear receptor element
- 3-AT
3-aminotriazole
- IPTG
isopropyl β-d-thiogalactoside
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.110149297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.110149297
References
- 1.Allen J B, Walberg M W, Edwards M C, Elledge S J. Trends Biol Sci. 1995;20:511–516. doi: 10.1016/s0968-0004(00)89119-7. [DOI] [PubMed] [Google Scholar]
- 2.Phizicky E M, Fields S. Microbiol Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rebar E J, Greisman H A, Pabo C O. Methods Enzymol. 1996;267:129–149. doi: 10.1016/s0076-6879(96)67010-4. [DOI] [PubMed] [Google Scholar]
- 4.Smith G P, Petrenko V A. Chem Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 5.Vidal M, Legrain P. Nucleic Acids Res. 1999;27:919–929. doi: 10.1093/nar/27.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolfe S A, Greisman H A, Ramm E I, Pabo C O. J Mol Biol. 1999;285:1917–1934. doi: 10.1006/jmbi.1998.2421. [DOI] [PubMed] [Google Scholar]
- 7.Hu J C, Kornacker M G, Hochschild A. Methods. 2000;20:80–94. doi: 10.1006/meth.1999.0908. [DOI] [PubMed] [Google Scholar]
- 8.Dove S L, Joung J K, Hochschild A. Nature (London) 1997;386:627–630. doi: 10.1038/386627a0. [DOI] [PubMed] [Google Scholar]
- 9.Kornacker M G, Remsburg B, Menzel R. Mol Microbiol. 1998;30:615–624. doi: 10.1046/j.1365-2958.1998.01096.x. [DOI] [PubMed] [Google Scholar]
- 10.Dove S L, Hochschild A. Genes Dev. 1998;12:745–754. doi: 10.1101/gad.12.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karimova G, Pidoux J, Ullmann A, Ladant D. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pelletier J N, Campbell-Valois F-X, Michnick S W. Proc Natl Acad Sci USA. 1998;95:12141–12146. doi: 10.1073/pnas.95.21.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greisman H A, Pabo C O. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 14.Farrell S, Simkovich N, Wu Y, Barberis A, Ptashne M. Genes Dev. 1996;10:2359–2367. doi: 10.1101/gad.10.18.2359. [DOI] [PubMed] [Google Scholar]
- 15.Whipple F W. Nucleic Acids Res. 1998;26:3700–3706. doi: 10.1093/nar/26.16.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfe, S. A., Ramm, E. I. & Pabo, C. O. (2000) Structure, in press. [DOI] [PubMed]
- 17.Struhl K, Cameron J R, Davis R W. Proc Natl Acad Sci USA. 1976;73:1471–1475. doi: 10.1073/pnas.73.5.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Struhl K, Davis R W. Proc Natl Acad Sci USA. 1977;74:5255–5259. doi: 10.1073/pnas.74.12.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brennan M B, Struhl K. J Mol Biol. 1980;136:333–338. doi: 10.1016/0022-2836(80)90377-0. [DOI] [PubMed] [Google Scholar]
- 20.Hollingshead J, Vapnek D. Plasmid. 1984;13:17–30. doi: 10.1016/0147-619x(85)90052-6. [DOI] [PubMed] [Google Scholar]
- 21.Wolfe S A, Nekludova L, Pabo C O. Annu Rev Biophys Biomol. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 22.Dove S L, Hochschild A. Cold Spring Harbor Symp Quant Biol. 1998;63:173–180. doi: 10.1101/sqb.1998.63.173. [DOI] [PubMed] [Google Scholar]
- 23.Fernandes P B. Curr Opin Chem Biol. 1998;2:597–603. doi: 10.1016/s1367-5931(98)80089-6. [DOI] [PubMed] [Google Scholar]