Abstract
Cytosine methylation is a characteristic property of prokaryotic and eukaryotic genomes. In the latter, it is indispensable for a healthy development of the organism and uncontrolled changes in the distribution of 5-methylcytosine (5mC) have been linked to severe disorders, in particular cancer. The growing scientific interest in DNA methylation has led to a considerable amount of data about this epigenetic phenomenon. In order to make these data readily available, we have established a dedicated database. The DNA Methylation database (MethDB) is currently the only public database for DNA methylation (http://www.methdb.net). This constantly growing database has become a key resource in the field of DNA methylation research. The database contains currently methylation patterns, profiles and total methylation content data for 46 species, 160 tissues and 72 phenotypes coming from a total of 6667 experiments (as of September 4, 2002). About 14% of the data have not been published elsewhere. These data can be conveniently searched and represented in different ways. Recently, we have included an on-line submission tool that permits the scientific public to directly enter new data into MethDB.
INTRODUCTION
Methylation of the 5′ position of the pyrimidine ring of cytosine residues is an epigenetic (i.e. heritable but potentially reversible) modification of the DNA in most organisms. In eukaryotes, DNA methylation patterns are relatively faithfully maintained from one cell generation to the other but many developmental changes are either induced or accompanied by dramatic changes in the methylation level of isolated loci or whole genomic regions. Among these, the global and local DNA methylation changes that are associated with tumorigenesis have attracted considerable scientific resources (reviewed in 1). DNA methylation is only one part of the epigenetic network that enables the organism to organise the genetic information contained in the sequence of the DNA molecule. And recently, many new insights have been gained about the interrelation of histone modifications, DNA methylation and noncoding RNA. However, 5-methylcytosine (5mC) remains the most easily analysable part in this network. While 5mC has been known for decades, the availability of new methods to analyse DNA methylation and the increasing awareness of the importance of epigenetic phenomena in normal and pathologic development of the organism has lead to an explosion of data in this field. This has prompted us to introduce the public DNA methylation database (MethDB) as a resource to store and to standardise DNA methylation data (http://www.methdb.net). Currently, the database contains methylation patterns, profiles and total methylation content data for 46 species, 160 tissues and 72 phenotypes coming from a total of 6667 experiments. The availability of such a database has met a warm welcome by the scientific community and MethDB is constantly gaining popularity. The number of monthly requests has increased from 810 in June 2000 to 8884 in July 2002. This encouraged us to further improve the database and we present here new features in MethDB. A guide how to search the database has been published earlier (2).
IMPROVEMENTS
Description of experimental conditions
A particularity of DNA methylation data is their heterogeneity. Methylation patterns of single DNA molecules go side by side with estimations of the 5mC content of whole genomes. On the other hand, a multitude of different methods have been developed that deliver results that are in principle comparable. For instance, a particular cytosine site could have been analysed by isoschizomeric digest (3), COBRA (4), Ms-SNuPE (5) or bisulfite genomic sequencing (6), all giving a relative methylation level at this site. However, each of these methods has its advantages and limitations. From the beginning, the policy of MethDB was therefore to indicate for each experiment the method that was used and to outline its principle. If PCR was used, it is now possible to display the exact amplification conditions and the primers that were used. Hybridisation probes are available under the ‘sequence’ link. This will aid the expert to evaluate the results, and makes it easier to repeat published experiments.
Graphical data representation
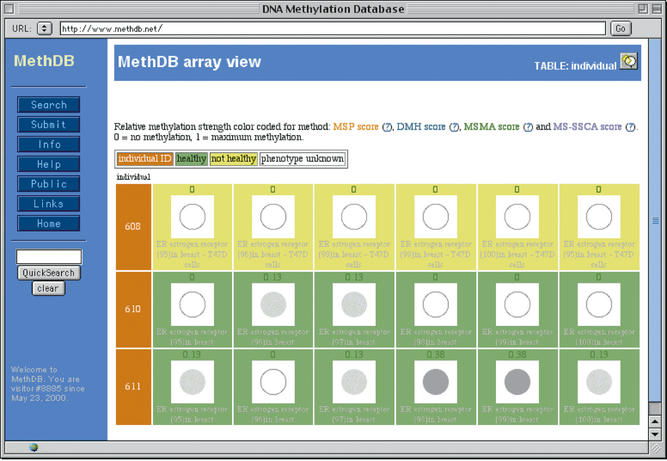
Methylation patterns (the 5mC/C/T/A/G sequence of a DNA molecule) and DNA profiles (the average methylation along a sequence) are dynamically displayed in a graphical way. These graphs are now client-side image maps and the details are available by clicking on the image, which we hope is more intuitive than the previous way. Methylation content data (i.e. the amount of 5mC in a set of DNA molecules) were so far only available as numerical values. We have now established a representation that displays these data as greyscales in an array like fashion (‘array view’, Fig. 1). This allows for rapid comparison of many individual experiments and mimics the results obtained by (micro)array approaches. Again, details are accessible by clicking on the image.
Figure 1.
Example of the representation of 5mC content data. Each row of the table corresponds to an investigated individual with its identification numbers (ID) in the first column. Each column corresponds to a specific locus that was analysed. 5-methylcytosine content is indicated on the top of each cell (in this case in relative units between 0 and 1). The greyscaled spot below the numeric value represents the 5mC content: from a hollow circle for 0 (no methylation) to a full black circle for 1 (maximum methylation). Each spot is hyperlinked to more detailed information about the experiment.
Phenotype descriptions
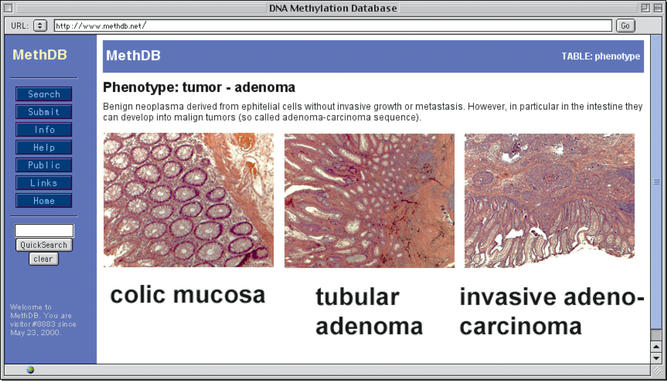
The particular interest of DNA methylation as an epigenetic mark lies in its association with a certain phenotype, developmental state and/or a tissue. We have, therefore, re-edited tissue and phenotype descriptions with an emphasis on tumour descriptions. MethDB does not attempt to be comprehensive in this regard, but delivers a short instructive description to somebody who is not an expert in this field. Links are provided to specialised on-line resources for further information. Tumour diagnosis is often based on histopathological evidence. We believe that for many users such microscopical images might not be readily available. We, therefore, gradually add annotated histopathological microphotographs to the database. Technically, this was done by adding a new table containing these images to MethDB, and creating relations to the tissue table and the phenotype table from there. An example output is shown in Figure 2.
Figure 2.
Example of a phenotype description with histopathological micrographs.
On-line submission tool
MethDB is a public database and users are invited to submit their own data. So far that had to be done by personal communication with the annotators. To facilitate and accelerate submission, we have now created on-line forms that allow for remote data submission (follow link ‘Submit’). New authors are demanded to supply their email address as login and information about how they can be contacted. Submitted data goes into a dedicated database and is subsequently verified by an annotator. In case of doubt the author is contacted. Fully annotated entries are regularly transferred to the read-only version of MethDB, and can then be searched via the query forms (follow link ‘Search’). As an alternative to the on-line form, data can still be submitted by email to submit@methdb.de. This is especially useful if the data are already available in an electronic form (e.g. a computer worksheets or local databases).
PROSPECTS
The focus of the work on MethDB will lie on the further development of the submission tool in order to make data entry as easy as possible. We also plan to establish versions of the database that are searchable via the Sequence Retrieval Systems (SRS) (http://www.lionbioscience.com/) and the Distributed Annotation System (DAS) (http://www.biodas.org/).
Acknowledgments
ACKNOWLEDGEMENTS
The work presented here was in part supported by a grant of the Klaus Tschira Foundation (Heidelberg, Germany) and the Genopole Languedoc-Roussillon (Montpellier, France). The authors are grateful to the scientists who have supplied supplementary information and unpublished data and to G. Roizes for carefully reading the manuscript. The concept of the database, query forms and output was designed by C.G., phenotype descriptions and micrographs were done by W.H., C.A. and C.G. have designed the submission tool.
REFERENCES
- 1.Costello J.F. and Plass,C. (2001) Methylation matters. J. Med. Genet., 38, 285–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grunau C., Renault,E. and Roizes,G. (2002) DNA Methylation Database ‘MethDB’: a user guide. J. Nutr., 132, 2435S–2439S. [DOI] [PubMed] [Google Scholar]
- 3.Bird A.P. and Southern,E.M. (1978) Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J. Mol. Biol., 118, 27–47. [DOI] [PubMed] [Google Scholar]
- 4.Xiong Z.G. and Laird,P.W. (1997) Cobra: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res., 25, 2532–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalgo M.L. and Jones,P.A. (1997) Rapid quantitation of methylation differences at specific sites using methylation-sensitive single nucleotide primer extension (Ms-SNuPE). Nucleic Acids Res., 25, 2529–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frommer M., McDonald,L.E., Millar,D.S., Collis,C.M., Watt,F., Grigg,G.W., Molloy,P.L. and Paul,C.L. (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA, 89, 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]