Abstract
The ProtoNet site provides an automatic hierarchical clustering of the SWISS-PROT protein database. The clustering is based on an all-against-all BLAST similarity search. The similarities' E-score is used to perform a continuous bottom-up clustering process by applying alternative rules for merging clusters. The outcome of this clustering process is a classification of the input proteins into a hierarchy of clusters of varying degrees of granularity. ProtoNet (version 1.3) is accessible in the form of an interactive web site at http://www.protonet.cs.huji.ac.il. ProtoNet provides navigation tools for monitoring the clustering process with a vertical and horizontal view. Each cluster at any level of the hierarchy is assigned with a statistical index, indicating the level of purity based on biological keywords such as those provided by SWISS-PROT and InterPro. ProtoNet can be used for function prediction, for defining superfamilies and subfamilies and for large-scale protein annotation purposes.
INTRODUCTION
Protein classification algorithms are roughly divided to those based on motif and domain analyses and those that rely on whole protein analysis (1). The latter have to deal with the problem that many proteins consist of several domains. Consequently, assuming that protein similarity is transitive may lead to classifying together non-related proteins that share some highly conserved domains (but not others).
The advantages of clustering proteins and the observation that it may provide insight based on transitivity have been studied and implemented in various systems, such as ProtoMap (2), Picasso (3), SYSTERS (4) and CluSTr (5).
Our clustering methods depend on a standard similarity measure, namely gapped BLAST. We rely on the notion of transitivity in order to perform a continuous process of clustering. As this process progresses, we discover larger clusters, making use of weaker similarities. In previous work (6), predetermined thresholds were used for constructing the hierarchy and that resulted in discrete, somewhat arbitrary, stages. A ‘cluster's purity’ is defined by optimizing the sensitivity-specificity measures of keywords that are associated with the proteins within. Systematic inspection of the clusters reveals that ‘purity’ is achieved at a range of stages along the hierarchical process (Vaaknin, Sasson and Linial, unpublished observation). Herein, we allowed the procedure of clustering to progress continuously, so the resulting clusters have different levels of granularity. Novel statistical and graphical tools allow evaluation of the cluster quality against major databases such as PDB (7) and InterPro (8).
METHODS
The basis of our clustering process is a sequence similarity measure for each pair of the 94 152 proteins in SWISS-PROT 39.15 (9). We use standard gapped BLAST based on BLOSUM62 and on filtration of low complexity sequences.
We encode the BLAST results through a graph whose vertex set is the collection of all SWISS-PROT proteins. Two vertices are connected by an edge if their pair-wise similarity exceeds a significance level corresponding to an E-score of 10. Our earlier experience shows (10,11) that even at that low BLAST score meaningful biological information is encoded.
Our clustering method is an adaptation of the hierarchical clustering paradigm. The clustering algorithm starts out with each protein as a singleton cluster. It then iteratively merges a pair of clusters, always selecting the pair whose merging has the lowest merging score. We use different merging scores, producing different clustering hierarchies. Recall that our clustering algorithm consists of a sequence of cluster merges. We seek to define a score for all possible merges so that the algorithm always carries out the currently most beneficial scoring merging step. The basic quantities on which our whole analysis is based are BLAST's pair-wise E-scores. Thus we wish to compare the average inter-cluster E-scores before and after a possible merge. There are numerous ways of averaging, and here we consider three averaging rules based on which we create three maps of the proteins space, based on Arithmetic, Geometric and Harmonic averages (for short referred to as A, G and H, respectively). A simple inequality ties all these averages. The H-mean is less than or equal to the G-mean which in turn is less than or equal to the A-mean. The rational and the performance of each of the averaging merging rules is discussed (12).
THE ProtoNet WEB-SITE
The ProtoNet (version 1.3) website is accessible at http://www.protonet.cs.huji.ac.il. The website allows easy navigation through the hierarchy of clusters created. This site offers a rich set of queries that allow the user to explore the protein space. This site is of interest to lab biologists who are interested in specific protein families and their inter-relations. It should also be a valuable tool for various types of research activities in computational biology, as ProtoNet clusters provide a global view on evolution and function of protein families.
The website provides a ‘guided tour’ to allow new users to learn about the different options as well as background information on the clustering process. Below we describe some of the main features and queries available to the user of the ProtoNet website. Additional updates are posted in ‘ProtoNet news’.
SEARCHING FOR PROTEINS
ProtoNet allows searching for proteins by their ProtoNet identifier, their SWISS-PROT accession number or ID (9) and by a protein's name (or any word contained in it) (Fig. 1).
Figure 1.
ProtoNet protein card.
The information displayed in the protein card for a single protein includes a condensed summary on each of the proteins in the database as extracted from SWISS-PROT information. This includes the SWISS-PROT ID and its accession number, update date and the full protein name (9). In addition, the protein length, PROSITE (13) and InterPro (8) accession numbers, and the PDB (7) identifiers (where applicable) are presented. Each protein is associated with an internal ProtoNet identification. Furthermore, the actual sequence is displayed and indicated by 7-colour coded amino-acid groups, as well as the protein's taxonomy.
To view the protein by its identified domains we provide a graphic display for motifs and domains according to the integration scheme used by InterPro (8) based on PROSITE (13), ProDom (14), Pfam (15), SMART (16) and Prints (17). The domains and motifs along the protein sequence and the sequence covered by any of the domains are also displayed.
The protein card allows one to move on to the corresponding cluster for each of the clustering algorithms (H, G and A). It is advised to investigate your protein of interest by comparing the performance of the different maps created. The navigation and comparative tools are described below.
NAVIGATING THE CLUSTER HIERARCHY
Vertical perspective
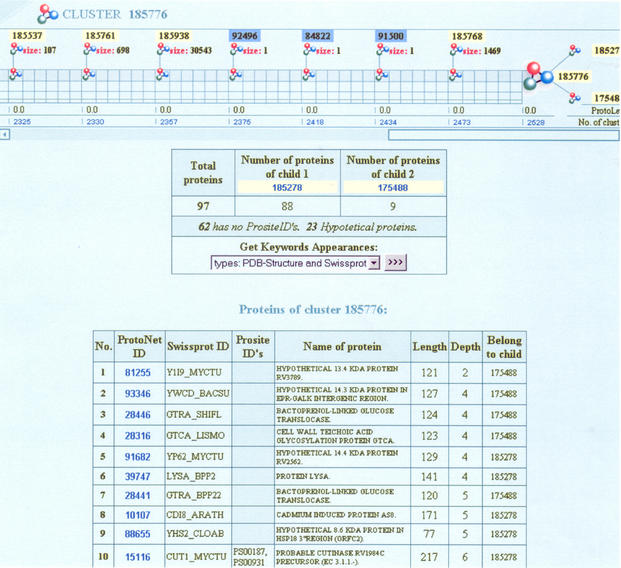
The cluster hierarchy is navigated using the cluster page. The cluster page provides an exhaustive description of the current cluster and the clusters from whose merger it was created. Specifically, it shows a breakdown of the number of proteins belonging to each of its two children (i.e. the two clusters merged to form this one), the number of proteins within the cluster to which no PROSITE (13) information is available as well as the number of hypothetical proteins and fragments that are included.
In addition, a table listing the proteins in the cluster and their association with each of the children is presented, As well as a tool for aligning any protein pair via BLAST.
The top of the cluster page provides a graphical navigation tool for browsing the cluster hierarchy at high resolution. This navigation pane shows the current cluster, the clusters whose merging created it, and the sequence of larger clusters generated from subsequent merges. For a detailed view of the process of merging, pop-up boxes are available. Information provided by these boxes includes the number of clusters at each merging step, the number of proteins that were not yet clustered at that level of the hierarchy (i.e. singletons, marked by Sg), the ProtoNet identifiers of the merging clusters and their appropriate size.
A cumulative list of keywords for each cluster is provided. A table showing the frequency of keywords from SWISS-PROT (9), PDB (7), InterPro (8) and taxonomy (18) provides an easy way to track the ‘purity’ of generated clusters. To further enhance the ability to track the cluster's ‘purity’ we included the breakdown of keywords of each of the clusters' children in comparison to the current cluster with statistical measures for each of the keywords.
Additional tables (marked by Cluster info) summarize relevant biological information for the cluster, such as the average length of proteins belonging to that cluster, the number of solved proteins according to PDB (7) and more. Features of the cluster under inspection in the context of the entire hierarchy are also included, such as the number of merging steps that were involved in creating that cluster and the number of clusters in this level of the hierarchy. These features can be used in comparing the global properties of the clusters for each of the merging procedures described.
Moving from a protein card to its cluster page and to external links: PDB (7), PROSITE (13), InterPro (8), SWISS-PROT keywords (9) and taxonomy (18) is conveniently done by links.
Horizontal perspective
ProtoNet allows exploration of the neighbourhood of a cluster. To this end, we include a list of neighbours according to their distance from the cluster of interest, and their size. The user can choose the preferred level in the hierarchy by defining the number of clusters (with or without singletons) or according to a selected ProtoLevel. ProtoLevel is a normalized measure for the progress of the clustering process and indicates the distance of a cluster to the root (ranging from 0–100). For example, ProtoLevel 30.0 for each of the algorithms H, G and A, respectively, consists of 4165, 6268 and 8401 clusters (and 10 249, 12 928 and 16 018 clusters including singletons). Most proteins are clustered to only few big components while 1508 clusters remains singletons even at the root level (Fig. 2).
Figure 2.
ProtoNet cluster page.
CLASSIFYING NEW PROTEINS
The issue of updating the clustering is an important one. Large numbers of new proteins are discovered, and it is of high interest to ‘map’ them into an existing clustering, since this can shed light on their structure and function.
ProtoNet implements an algorithm for mapping a new protein into an existing clustering system, which is based on performing a BLAST computation of the new protein against all sequences stored in the database. The clustering for the new protein is approximated using the protein closest to this new sequence in the BLAST similarity test (to be described elsewhere). Practically, a user may classify a new sequence and trace its merging within the current ProtoNet graph.
Updating the database and including proteins from SWISS-PROT (9) updates will be carried out once a year. The older versions will be accessible to allow consistency with publications based on a specific ProtoNet release.
NAVIGATION AIDS
In order to ease the task of navigating the clustering system, ProtoNet provides a powerful history mechanism that allows the user to return to any of the most recently visited proteins and clusters (20 steps). Each cluster that was visited before is associated with the merging rules used (G, H or A).
CONNECTIONS WITHIN AND AMONG CLUSTERS
ProtoNet provides several mechanisms for studying the resulting clusters. In addition to the details provided in the cluster page, it is possible to retrieve the breakdown of sequence similarity for two clusters. This breakdown consists of sequence similarities for all protein pairs within each of the clusters and for all proteins pairs composed of one protein from the first cluster and another member from the other cluster. The distribution of sequence similarities shows how strongly two clusters are related, and has a direct impact on the likelihood of two clusters merging. Furthermore, the user may enquire for the smallest cluster containing two given proteins. This last query is accessed from ‘Get Cluster Card’ option.
CONCLUSIONS
ProtoNet provides a rich set of tools for comprehensive analysis of all protein sequences in the SWISS-PROT database (9). ProtoNet allows a dynamic view of protein clusters at different levels of granularity and according to varying merging rules. When studying a certain protein family, analyzing clusters according to the different merging rules is beneficial. The levels of purity of several known protein families were reported (12). The ability to classify a new sequence allows users to study their own sequences against the various clustering systems. Most importantly, the system facilitates the study of protein families, by providing the means to navigate the cluster hierarchy focusing on a desirable level of resolution. The statistics for each cluster included a measure for the false positive and false negative according to information obtained from SWISS-PROT keywords (9), InterPro annotations (8), PDB assignments (7) and more. These measures provide accurate and efficient means to select the desired hierarchical level.
Acknowledgments
ACKNOWLEDGEMENTS
This study could not be done without the contribution of our undergraduate students Shmuel Brody and Noam Kaplan. We thank Alexander Savenok for his excellent work in designing the website and Edna Wigderson and Hagit Mor for suggestions and fruitful discussions.
This work was supported by the Israeli Ministry of Defense, the Israeli Ministry of Science and by the Horowitz Foundation.
REFERENCES
- 1.Kriventseva E.V., Biswas,M. and Apweiler,R. (2001) Clustering and analysis of protein families. Curr. Opin. Struct. Biol., 11, 334–339. [DOI] [PubMed] [Google Scholar]
- 2.Yona G., Linial,N. and Linial,M. (2000) ProtoMap: Automatic classification of protein sequences and hierarchy of protein families. Nucleic Acids Res., 28, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heger A. and Holm,L. (2001) Picasso: generating a covering set of protein family profiles. Bioinformatics, 17, 272–279. [DOI] [PubMed] [Google Scholar]
- 4.Krause A., Stoye,J. and Vingron,M. (2000) The SYSTERS protein sequence cluster set. Nucleic Acids Res., 28, 270–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kriventseva E.V., Fleischmann,W., Zdobnov,E.M. and Apweiler,R. (2001) CluSTr: a database of clusters of SWISS-PROT+TrEMBL proteins. Nucleic Acids Res., 29, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yona G., Linial,N. and Linial,M. (1999) ProtoMap—Automated classification of all proteins sequences: a hierarchy of protein families, and local maps of the protein space. Proteins, 37, 360–378. [PubMed] [Google Scholar]
- 7.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Apweiler R., Attwood,T.K., Bairoch,A., Bateman,A., Birney,E., Biswas,M., Bucher,P., Cerutti,L., Corpet,F., Croning,M.D.R. et al. (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res., 29, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Portugaly E. and Linial,M. (2000) Estimating the probability for a protein to have a new fold: A statistical computational model. Proc. Natl Acad. Sci. USA, 97, 5161–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Portugaly E., Kifer,I. and Linial,M. (2002) Selecting targets for structural determination by navigating in a graph of protein families. Bioinformatics, 18, 899–907. [DOI] [PubMed] [Google Scholar]
- 12.Sasson O., Linial,N. and Linial,M. (2002) The metric space of proteins—comparative study of clustering algorithms. Bioinformatics, 18, S14–S21. [DOI] [PubMed] [Google Scholar]
- 13.Hofmann K., Bucher,P., Falquet,L. and Bairoch,A. (1999) The PROSITE database, its status in 1999. Nucleic Acids Res., 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corpet F., Servant,F., Gouzy,J. and Kahn,D. (2000) ProDom and ProDom-CG: tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res., 28, 267–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Howe,K.L. and Sonnhammer,E.L. (2000) The Pfam protein families database. Nucleic Acids Res., 28, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultz J., Copley,R.R., Doerks,T., Ponting,C.P. and Bork,P. (2000) SMART: A Web-based tool for the study of genetically mobile domains. Nucleic Acids Res., 28, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Attwood T.K., Croning,M.D.R., Flower,D.R., Lewis,A.P., Mabey,J.E., Scordis,P., Selley,J.N. and Wright,W. (2000) PRINTS-S: the database formerly known as PRINTS. Nucleic Acids Res., 28, 225–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheeler D.L., Chappey,C., Lash,A.E., Leipe,D.D., Madden,T.L., Schuler,G.D., Tatusova,T.A. and Rapp,B.A. (2000) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 28, 10–14. [DOI] [PMC free article] [PubMed] [Google Scholar]